|
Molecular Tools For Gene Study
Nucleic acid methods are the techniques used to study nucleic acids: DNA and RNA. Purification *DNA extraction * Phenol–chloroform extraction * Minicolumn purification *RNA extraction *Boom method *Synchronous coefficient of drag alteration (SCODA) DNA purification Quantification *Abundance in weight: spectroscopic nucleic acid quantitation *Absolute abundance in number: real-time polymerase chain reaction (quantitative PCR) *High-throughput relative abundance: DNA microarray *High-throughput absolute abundance: serial analysis of gene expression (SAGE) *Size: gel electrophoresis Synthesis *''De novo'': oligonucleotide synthesis *Amplification: polymerase chain reaction (PCR) Kinetics * Multi-parametric surface plasmon resonance *Dual-polarization interferometry * Quartz crystal microbalance with dissipation monitoring (QCM-D) Gene function *RNA interference Other * Bisulfite sequencing * DNA sequencing *Expression cloning *Fluorescence in situ hybridization *Lab-on-a-chip * ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid
Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomers made of three components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). If the sugar is ribose, the polymer is RNA; if the sugar is the ribose derivative deoxyribose, the polymer is DNA. Nucleic acids are naturally occurring chemical compounds that serve as the primary information-carrying molecules in cells and make up the genetic material. Nucleic acids are found in abundance in all living things, where they create, encode, and then store information of every living cell of every life-form on Earth. In turn, they function to transmit and express that information inside and outside the cell nucleus to the interior operations of the cell and ultimately to the next generation of each living organism. The encoded informatio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including ''co-suppression'', ''post-transcriptional gene silencing'' (PTGS), and ''quelling''. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm ''Caenorhabditis elegans'', which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatics
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combines biology, chemistry, physics, computer science, information engineering, mathematics and statistics to analyze and interpret the biological data. Bioinformatics has been used for '' in silico'' analyses of biological queries using computational and statistical techniques. Bioinformatics includes biological studies that use computer programming as part of their methodology, as well as specific analysis "pipelines" that are repeatedly used, particularly in the field of genomics. Common uses of bioinformatics include the identification of candidates genes and single nucleotide polymorphisms ( SNPs). Often, such identification is made with the aim to better understand the genetic basis of disease, unique adaptations, desirable propertie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Toeprinting Assay
The toeprinting assay, also known as the primer extension inhibition assay, is a method used in molecular biology that allows one to examine the interactions between messenger RNA and ribosomes or RNA-binding proteins. It is different from the more commonly used DNA footprinting assay. The toeprinting assay has been utilized to examine the formation of the translation initiation complex. To do a toeprint assay, one needs the mRNA of interest, ribosomes, a DNA primer, free nucleotides, and reverse transcriptase (RT), among other reagents. The assay involves letting the RT generate cDNA In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA (e.g., messenger RNA (mRNA) or microRNA (miRNA)) template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to express a speci ... until it gets blocked by any bound ribosomes, resulting in shorter fragments called toeprints when the results are observed on a sequencing gel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Differential Centrifugation
In biochemistry and cell biology, differential centrifugation (also known as differential velocity centrifugation) is a common procedure used to separate organelles and other sub-cellular particles based on their sedimentation rate. Although often applied in biological analysis, differential centrifugation is a general technique also suitable for crude purification of non-living suspended particles (e.g. nanoparticles, colloidal particles, viruses). In a typical case where differential centrifugation is used to analyze cell-biological phenomena (e.g. organelle distribution), a tissue sample is first lysed to break the cell membranes and release the organelles and cytosol. The lysate is then subjected to repeated centrifugations, where particles that sediment sufficiently quickly at a given centrifugal force for a given time form a compact "pellet" at the bottom of the centrifugation tube. After each centrifugation, the ''supernatant'' (non-pelleted solution) is removed from t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Southern Blot
A Southern blot is a method used in molecular biology for detection of a specific DNA sequence in DNA samples. Southern blotting combines transfer of electrophoresis-separated DNA fragments to a filter membrane and subsequent fragment detection by probe hybridization. The method is named after the British biologist Edwin Southern, who first published it in 1975. Other blotting methods (i.e., western blot, northern blot, eastern blot, southwestern blot) that employ similar principles, but using RNA or protein, have later been named in reference to Edwin Southern's name. As the label is eponymous, Southern is capitalised, as is conventional of proper nouns. The names for other blotting methods may follow this convention, by analogy. Method #Restriction endonucleases are used to cut high-molecular-weight DNA strands into smaller fragments. #The DNA fragments are then electrophoresed on an agarose gel to separate them by size. # If some of the DNA fragments are larg ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Radioactivity In The Life Sciences
Radioactivity is generally used in life sciences for highly sensitive and direct measurements of biological phenomena, and for visualizing the location of biomolecules radiolabelled with a radioisotope. All atoms exist as stable or unstable isotopes and the latter decay at a given half-life ranging from attoseconds to billions of years; radioisotopes useful to biological and experimental systems have half-lives ranging from minutes to months. In the case of the hydrogen isotope tritium (half-life = 12.3 years) and carbon-14 (half-life = 5,730 years), these isotopes derive their importance from all organic life containing hydrogen and carbon and therefore can be used to study countless living processes, reactions, and phenomena. Most short lived isotopes are produced in cyclotrons, linear particle accelerators, or nuclear reactors and their relatively short half-lives give them high maximum theoretical specific activities which is useful for detection in biological systems. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Run-on
A nuclear run-on assay is conducted to identify the genes that are being transcribed at a certain time point. Approximately one million cell nuclei are isolated and incubated with labeled nucleotides, and genes in the process of being transcribed are detected by hybridization of extracted RNA to gene specific probes on a blot. Garcia-Martinez et al. (2004) developed a protocol for the yeast ''S. cerevisiae'' (Genomic run-on, GRO) that allows for the calculation of transcription rates (TRs) for all yeast genes to estimate mRNA stabilities for all yeast mRNAs. Alternative microarray methods have recently been developed, mainly PolII RIP-chip: RNA immunoprecipitation of RNA polymerase II with phosphorylated C-terminal domain directed antibodies and hybridization on a microarray slide or chip (the word chip in the name stems from "ChIP-chip" where a special Affymetrix Affymetrix is now Applied Biosystems, a brand of DNA microarray products sold by Thermo Fisher Scientific that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
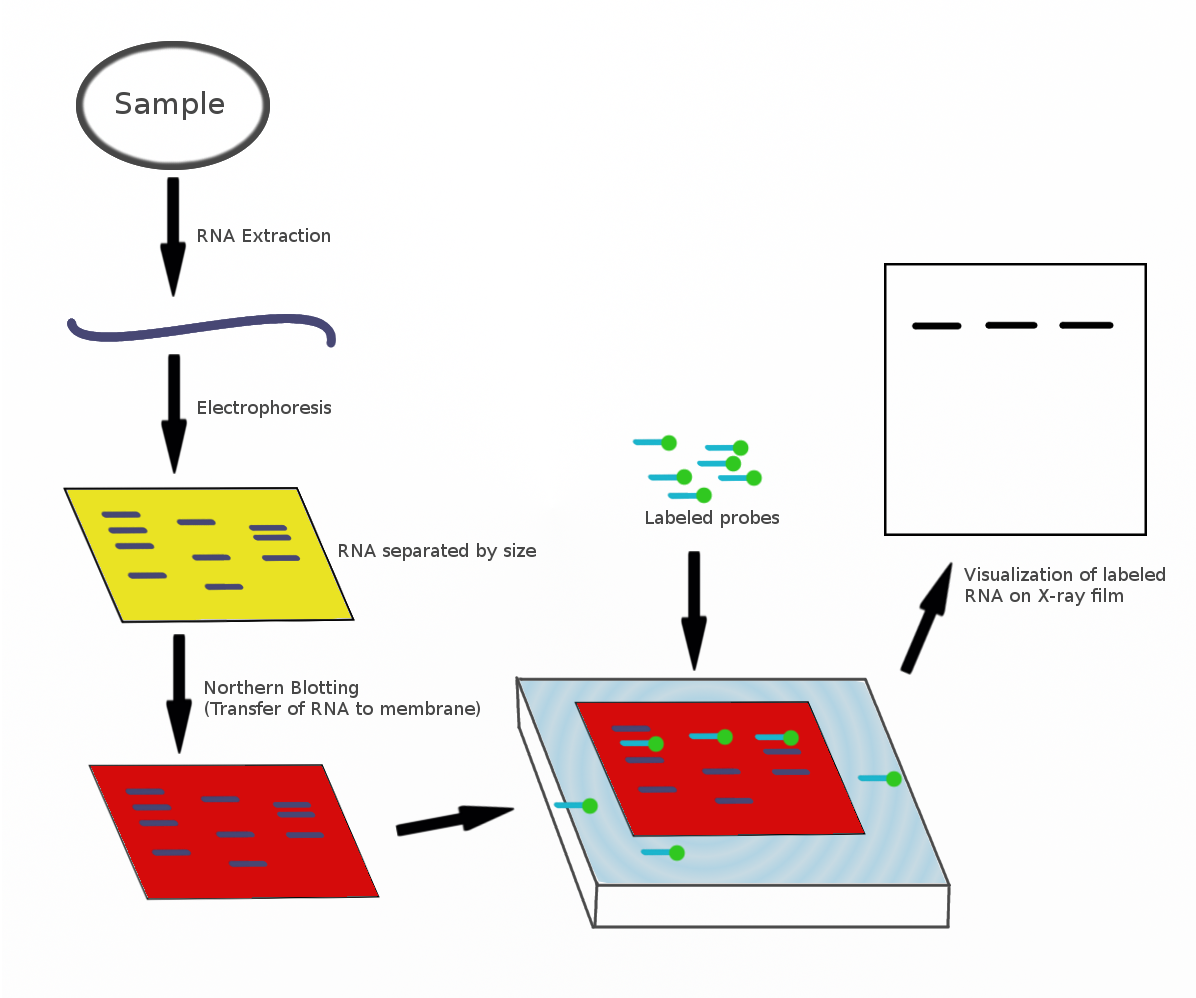
Northern Blot
The northern blot, or RNA blot,Gilbert, S. F. (2000) Developmental Biology, 6th Ed. Sunderland MA, Sinauer Associates. is a technique used in molecular biology research to study gene expression by detection of RNA (or isolated mRNA) in a sample.Kevil, C. G., Walsh, L., Laroux, F. S., Kalogeris, T., Grisham, M. B., Alexander, J. S. (1997) An Improved, Rapid Northern Protocol. Biochem. and Biophys. Research Comm. 238:277–279. With northern blotting it is possible to observe cellular control over structure and function by determining the particular gene expression rates during differentiation and morphogenesis, as well as in abnormal or diseased conditions. Northern blotting involves the use of electrophoresis to separate RNA samples by size, and detection with a hybridization probe complementary to part of or the entire target sequence. Strictly speaking, the term 'northern blot' refers specifically to the capillary transfer of RNA from the electrophoresis gel to the blotting m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Comparison Of Nucleic Acid Simulation Software
This is a list of notable computer programs that are used for nucleic acid Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomers made of three components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main ...s simulations. See also References Computational chemistry software Software comparisons Molecular dynamics software Molecular modelling software {{science-software-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lab-on-a-chip
A lab-on-a-chip (LOC) is a device that integrates one or several laboratory functions on a single integrated circuit (commonly called a "chip") of only millimeters to a few square centimeters to achieve automation and high-throughput screening. LOCs can handle extremely small fluid volumes down to less than pico-liters. Lab-on-a-chip devices are a subset of microelectromechanical systems (MEMS) devices and sometimes called "micro total analysis systems" (µTAS). LOCs may use microfluidics, the physics, manipulation and study of minute amounts of fluids. However, strictly regarded "lab-on-a-chip" indicates generally the scaling of single or multiple lab processes down to chip-format, whereas "µTAS" is dedicated to the integration of the total sequence of lab processes to perform chemical analysis. The term "lab-on-a-chip" was introduced when it turned out that µTAS technologies were applicable for more than only analysis purposes. History After the invention of microtechno ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fluorescence In Situ Hybridization
Fluorescence ''in situ'' hybridization (FISH) is a molecular cytogenetic technique that uses fluorescent probes that bind to only particular parts of a nucleic acid sequence with a high degree of sequence complementarity. It was developed by biomedical researchers in the early 1980s to detect and localize the presence or absence of specific DNA sequences on chromosomes. Fluorescence microscopy can be used to find out where the fluorescent probe is bound to the chromosomes. FISH is often used for finding specific features in DNA for use in genetic counseling, medicine, and species identification. FISH can also be used to detect and localize specific RNA targets (mRNA, lncRNA and miRNA) in cells, circulating tumor cells, and tissue samples. In this context, it can help define the spatial-temporal patterns of gene expression within cells and tissues. Probes – RNA and DNA In biology, a probe is a single strand of DNA or RNA that is complementary to a nucleotide sequence ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |

.jpg)