|
M13 Phage
M13 is one of the Ff phages (fd and f1 are others), a member of the family filamentous bacteriophage ( inovirus). Ff phages are composed of circular single-stranded DNA ( ssDNA), which in the case of the m13 phage is 6407 nucleotides long and is encapsulated in approximately 2700 copies of the major coat protein p8, and capped with about 5 copies each of four different minor coat proteins (p3 and p6 at one end and p7 and p9 at the other end). The minor coat protein p3 attaches to the receptor at the tip of the F pilus of the host ''Escherichia coli''. The life cycle is relatively short, with the early phage progeny exiting the cell ten minutes after infection. Ff phages are chronic phage, releasing their progeny without killing the host cells. The infection causes turbid plaques in ''E. coli'' lawns, of intermediate opacity in comparison to regular lysis plaques. However, a decrease in the rate of cell growth is seen in the infected cells. M13 plasmids are used for many recombi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ff Phages
Ff phages (for ''F'' specific ''f''ilamentous phages) is a group of almost identical filamentous phage (genus ''Inovirus'') including phages f1, fd, M13 and ZJ/2, which infect bacteria bearing the F fertility factor. The virion (virus particle) is a flexible filament measuring about 6 by 900 nm, comprising a cylindrical protein tube protecting a single-stranded circular DNA molecule at its core. The phage codes for only 11 gene products, and is one of the simplest viruses known. It has been widely used to study fundamental aspects of molecular biology. George Smith and Greg Winter used f1 and fd for their work on phage display for which they were awarded a share of the 2018 Nobel Prize in Chemistry. Early experiments on Ff phages used M13 to identify gene functions, and M13 was also developed as a cloning vehicle, so the name M13 is sometimes used as an informal synonym for the whole group of Ff phages. Structure The virion is a flexible filament (worm-like chain) about 6&nb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences (see non-coding DNA), and often a substantial fraction of 'junk' DNA with no evident function. Almost all eukaryotes have mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplasts with a chloroplast genome. The study of the genome is called genomics. The genomes of many organisms have been sequenced and various regions have been annotated. The International Human Genome Project reported the sequence of the genome for ''Homo sapiens'' in 200The Human Genome Project although the initial "finished" sequence was missing 8% of the genome consisting mostly of repetitive sequences. With advancements in technology that could handle sequenci ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phagemid
A phagemid or phasmid is a DNA-based cloning vector, which has both bacteriophage and plasmid properties. These vectors carry, in addition to the origin of plasmid replication, an origin of replication derived from bacteriophage. Unlike commonly used plasmids, phagemid vectors differ by having the ability to be packaged into the capsid of a bacteriophage, due to their having a genetic sequence that signals for packaging. Phagemids are used in a variety of biotechnology applications; for example, they can be used in a molecular biology technique called "Phage Display". Properties of the cloning vector A phagemid (plasmid + phage) is a plasmid that contains an f1 origin of replication from an f1 phage.Analysis of Genes and Genomes, John Wiley & Sons, 2004, S. 140Google Books/ref> It can be used as a type of cloning vector in combination with filamentous phage M13. A phagemid can be replicated as a plasmid, and also be packaged as single stranded DNA in viral particles. Phagemids ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phage Display
Phage display is a laboratory technique for the study of protein–protein, protein–peptide, and protein– DNA interactions that uses bacteriophages (viruses that infect bacteria) to connect proteins with the genetic information that encodes them. In this technique, a gene encoding a protein of interest is inserted into a phage coat protein gene, causing the phage to "display" the protein on its outside while containing the gene for the protein on its inside, resulting in a connection between genotype and phenotype. These displaying phages can then be screened against other proteins, peptides or DNA sequences, in order to detect interaction between the displayed protein and those other molecules. In this way, large libraries of proteins can be screened and amplified in a process called ''in vitro'' selection, which is analogous to natural selection. The most common bacteriophages used in phage display are M13 and fd filamentous phage, though T4, T7, and λ phage have ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endonuclease
Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (without regard to sequence), while many, typically called restriction endonucleases or restriction enzymes, cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (endo) portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity. Restriction enzymes are endonucleases from eubacteria and archaea that recognize a specific DNA sequence. The nucleotide sequence recognized for cleavage by a restriction enzyme is called the restriction site. Typically, a restriction site will be a palindromic sequence about four to six nucleotides ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Annual Reviews (publisher)
Annual Reviews is an independent, non-profit academic publishing company based in San Mateo, California. As of 2021, it publishes 51 journals of review articles and ''Knowable Magazine'', covering the fields of life, biomedical, physical, and social sciences. Review articles are usually “peer-invited” solicited submissions, often planned one to two years in advance, which go through a peer-review process. The organizational structure has three levels: a volunteer board of directors, editorial committees of experts for each journal, and paid employees. Annual Reviews' stated mission is to synthesize and integrate knowledge "for the progress of science and the benefit of society". The first Annual Reviews journal, the ''Annual Review of Biochemistry'', was published in 1932 under the editorship of Stanford University chemist J. Murray Luck, who wanted to create a resource that provided critical reviews on contemporary research. The second journal was added in 1939. By ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Liposome
A liposome is a small artificial vesicle, spherical in shape, having at least one lipid bilayer. Due to their hydrophobicity and/or hydrophilicity, biocompatibility, particle size and many other properties, liposomes can be used as drug delivery vehicles for administration of pharmaceutical drugs and nutrients, such as lipid nanoparticles in mRNA vaccines, and DNA vaccines. Liposomes can be prepared by disrupting biological membranes (such as by sonication). Liposomes are most often composed of phospholipids, especially phosphatidylcholine and cholesterol, but may also include other lipids, such as egg, phosphatidylethanolamine, as long as they are compatible with lipid bilayer structure. A liposome design may employ surface ligands for attaching to unhealthy tissue. The major types of liposomes are the multilamellar vesicle (MLV, with several lamellar phase lipid bilayers), the small unilamellar liposome vesicle (SUV, with one lipid bilayer), the large unilamellar vesicle ( ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Supercoil
DNA supercoiling refers to the amount of twist in a particular DNA strand, which determines the amount of strain on it. A given strand may be "positively supercoiled" or "negatively supercoiled" (more or less tightly wound). The amount of a strand’s supercoiling affects a number of biological processes, such as compacting DNA and regulating access to the genetic code (which strongly affects DNA metabolism and possibly gene expression). Certain enzymes, such as topoisomerases, change the amount of DNA supercoiling to facilitate functions such as DNA replication and transcription. The amount of supercoiling in a given strand is described by a mathematical formula that compares it to a reference state known as "relaxed B-form" DNA. Overview In a "relaxed" double-helical segment of B-DNA, the two strands twist around the helical axis once every 10.4–10.5 base pairs of sequence. Adding or subtracting twists, as some enzymes do, imposes strain. If a DNA segment under twist s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Type II Topoisomerase
Type II topoisomerases are topoisomerases that cut both strands of the DNA helix simultaneously in order to manage DNA tangles and supercoils. They use the hydrolysis of ATP, unlike Type I topoisomerase. In this process, these enzymes change the linking number of circular DNA by ±2. Topoisomerases are ubiquitous enzymes, found in all living organisms. In animals, topoisomerase II is a chemotherapy target. In prokaryotes, gyrase is an antibacterial target. Indeed, these enzymes are of interest for a wide range of effects. Function Type II topoisomerases increase or decrease the linking number of a DNA loop by 2 units, and it promotes chromosome disentanglement. For example, DNA gyrase, a type II topoisomerase observed in '' E. coli'' and most other prokaryotes, introduces negative supercoils and decreases the linking number by 2. Gyrase is also able to remove knots from the bacterial chromosome. Along with gyrase, most prokaryotes also contain a second type IIA topoisomerase, t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
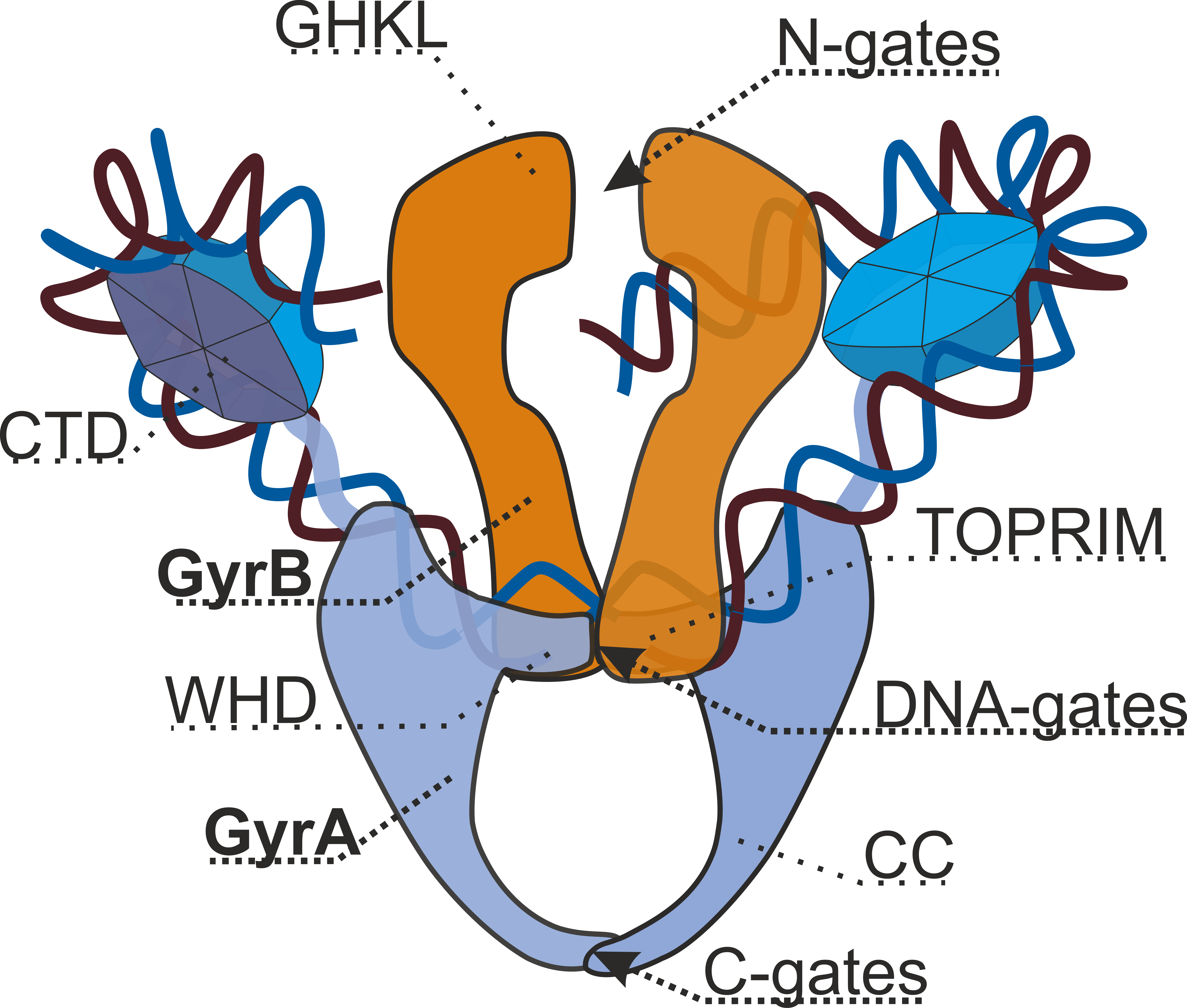
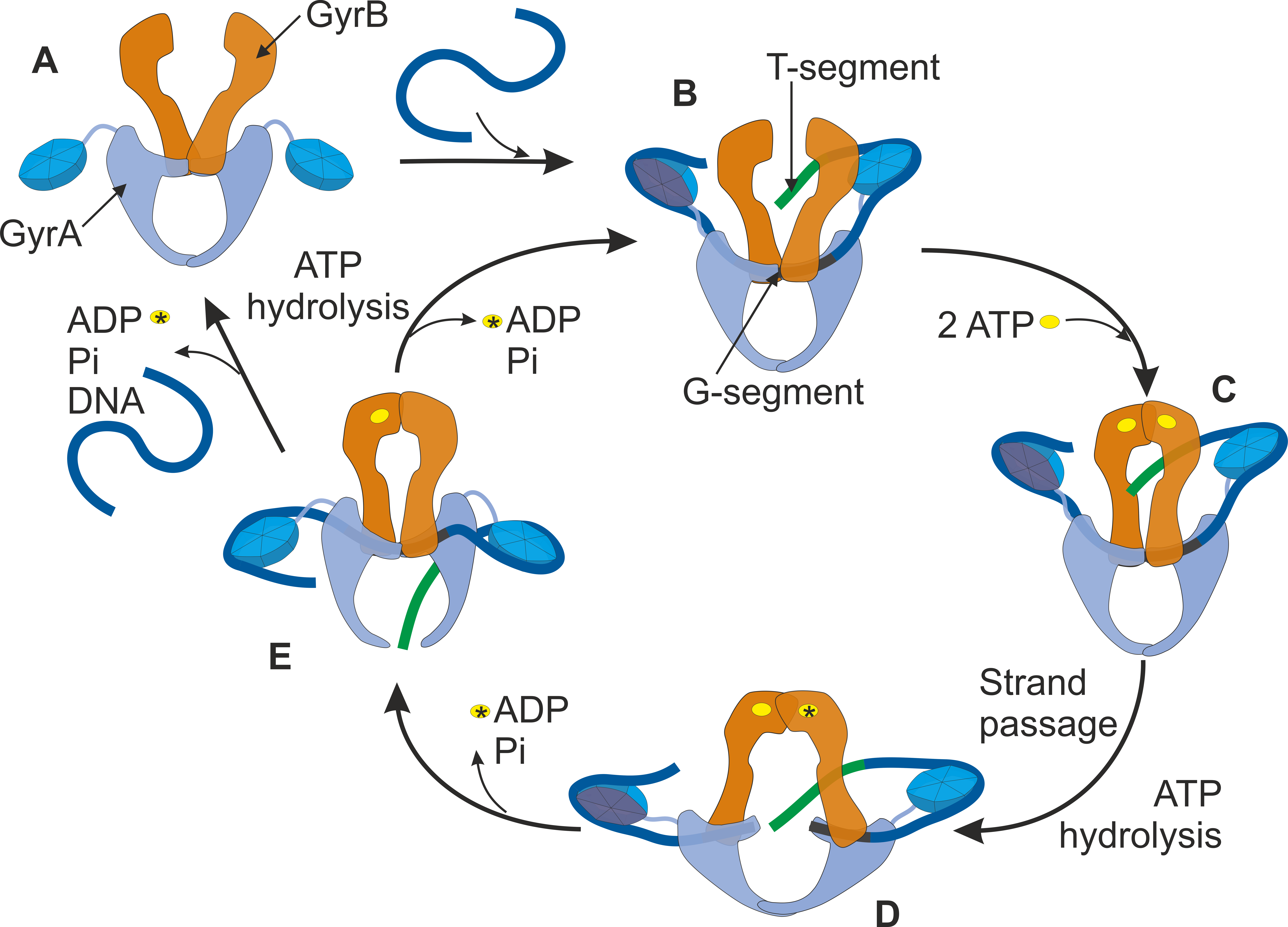
DNA Gyrase
DNA gyrase, or simply gyrase, is an enzyme Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products ... within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA polymerase, RNA-polymerase or by helicase in front of the progressing DNA replication#Replication fork, replication fork. The enzyme causes negative DNA supercoil, supercoiling of the DNA or relaxes positive supercoils. It does so by looping the template so as to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Complementary DNA
In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA (e.g., messenger RNA (mRNA) or microRNA (miRNA)) template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to express a specific protein in a cell that does not normally express that protein (i.e., heterologous expression), or to sequence or quantify mRNA molecules using DNA based methods (qPCR, RNA-seq). cDNA that codes for a specific protein can be transferred to a recipient cell for expression, often bacterial or yeast expression systems. cDNA is also generated to analyze transcriptomic profiles in bulk tissue, single cells, or single nuclei in assays such as microarrays, qPCR, and RNA-seq. cDNA is also produced naturally by retroviruses (such as HIV-1, HIV-2, simian immunodeficiency virus, etc.) and then integrated into the host's genome, where it creates a provirus. The term ''cDNA'' is also used, typically in a bioinformatics context, to refer to a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |