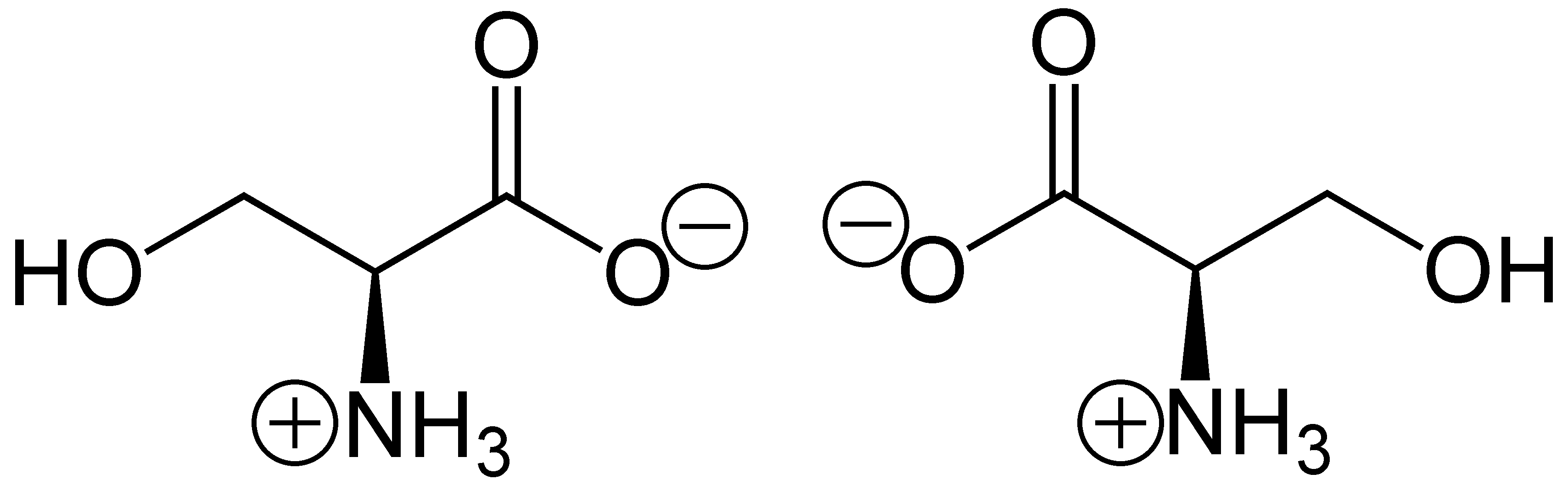|
L-serine Ammonia-lyase
The enzyme L-serine ammonia-lyase (EC 4.3.1.17) catalyzes the chemical reaction :L-serine = pyruvate + NH3 (overall reaction) ::(1a) L-serine = 2-aminoprop-2-enoate + H2O ::(1b) 2-aminoprop-2-enoate = 2-iminopropanoate (spontaneous) ::(1c) 2-iminopropanoate + H2O = pyruvate + NH3 (spontaneous) This enzyme belongs to the family of lyases, specifically ammonia lyases, which cleave carbon-nitrogen bonds. The systematic name of this enzyme class is L-serine ammonia-lyase (pyruvate-forming). Other names in common use include serine deaminase, L-hydroxyaminoacid dehydratase, L-serine deaminase, L-serine dehydratase, and L-serine hydro-lyase (deaminating). This enzyme participates in glycine, serine, threonine and cysteine metabolism. It employs one cofactor, pyridoxal phosphate. Structural studies As of late 2007, 4 structures A structure is an arrangement and organization of interrelated elements in a material object or system, or the object or system so organized. Material ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Catalysis
Catalysis () is the process of increasing the rate of a chemical reaction by adding a substance known as a catalyst (). Catalysts are not consumed in the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recycles quickly, very small amounts of catalyst often suffice; mixing, surface area, and temperature are important factors in reaction rate. Catalysts generally react with one or more reactants to form intermediates that subsequently give the final reaction product, in the process of regenerating the catalyst. Catalysis may be classified as either homogeneous, whose components are dispersed in the same phase (usually gaseous or liquid) as the reactant, or heterogeneous, whose components are not in the same phase. Enzymes and other biocatalysts are often considered as a third category. Catalysis is ubiquitous in chemical industry of all kinds. Estimates are that 90% of all commercially produced chemical products involve catalysts at some s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemical Reaction
A chemical reaction is a process that leads to the IUPAC nomenclature for organic transformations, chemical transformation of one set of chemical substances to another. Classically, chemical reactions encompass changes that only involve the positions of electrons in the forming and breaking of chemical bonds between atoms, with no change to the Atomic nucleus, nuclei (no change to the elements present), and can often be described by a chemical equation. Nuclear chemistry is a sub-discipline of chemistry that involves the chemical reactions of unstable and radioactive Chemical element, elements where both electronic and nuclear changes can occur. The substance (or substances) initially involved in a chemical reaction are called reagent, reactants or reagents. Chemical reactions are usually characterized by a chemical change, and they yield one or more Product (chemistry), products, which usually have properties different from the reactants. Reactions often consist of a sequence o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lyase
In biochemistry, a lyase is an enzyme that catalyzes the breaking (an elimination reaction) of various chemical bonds by means other than hydrolysis (a substitution reaction) and oxidation, often forming a new double bond or a new ring structure. The reverse reaction is also possible (called a Michael reaction). For example, an enzyme that catalyzed this reaction would be a lyase: : ATP → cAMP + PPi Lyases differ from other enzymes in that they require only one substrate for the reaction in one direction, but two substrates for the reverse reaction. Nomenclature Systematic names are formed as "''substrate group-lyase''." Common names include decarboxylase, dehydratase, aldolase, etc. When the product is more important, synthase may be used in the name, e.g. phosphosulfolactate synthase (EC 4.4.1.19, Michael addition of sulfite to phosphoenolpyruvate). A combination of both an elimination and a Michael addition is seen in O-succinylhomoserine (thiol)-lyase (MetY or MetZ) which ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Enzymes
This article lists enzymes by their classification in the International Union of Biochemistry and Molecular Biology's Enzyme Commission (EC) numbering system. * List of EC numbers (EC 5) * List of EC numbers (EC 6) :Oxidoreductases (EC 1) (Oxidoreductase) *Dehydrogenase * Luciferase *DMSO reductase :EC 1.1 (act on the CH-OH group of donors) * :EC 1.1.1 (with NAD+ or NADP+ as acceptor) ** Alcohol dehydrogenase (NAD) ** Alcohol dehydrogenase (NADP) **Homoserine dehydrogenase ** Aminopropanol oxidoreductase **Diacetyl reductase **Glycerol dehydrogenase **Propanediol-phosphate dehydrogenase ** glycerol-3-phosphate dehydrogenase (NAD+) ** D-xylulose reductase **L-xylulose reductase **Lactate dehydrogenase **Malate dehydrogenase **Isocitrate dehydrogenase ** HMG-CoA reductase * :EC 1.1.2 (with a cytochrome as acceptor) * :EC 1.1.3 (with oxygen as acceptor) **Glucose oxidase **L-gulonolactone oxidase **Thiamine oxidase **Xanthine oxidase * :EC 1.1.4 (with a disul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glycine Metabolism
Glycine (symbol Gly or G; ) is an amino acid that has a single hydrogen atom as its side chain. It is the simplest stable amino acid (carbamic acid is unstable), with the chemical formula NH2‐ CH2‐ COOH. Glycine is one of the proteinogenic amino acids. It is encoded by all the codons starting with GG (GGU, GGC, GGA, GGG). Glycine is integral to the formation of alpha-helices in secondary protein structure due to its compact form. For the same reason, it is the most abundant amino acid in collagen triple-helices. Glycine is also an inhibitory neurotransmitter – interference with its release within the spinal cord (such as during a ''Clostridium tetani'' infection) can cause spastic paralysis due to uninhibited muscle contraction. It is the only achiral proteinogenic amino acid. It can fit into hydrophilic or hydrophobic environments, due to its minimal side chain of only one hydrogen atom. History and etymology Glycine was discovered in 1820 by the French chemist Henr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine Metabolism
Serine (symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − form under biological conditions), and a side chain consisting of a hydroxymethyl group, classifying it as a polar amino acid. It can be synthesized in the human body under normal physiological circumstances, making it a nonessential amino acid. It is encoded by the codons UCU, UCC, UCA, UCG, AGU and AGC. Occurrence This compound is one of the naturally occurring proteinogenic amino acids. Only the L-stereoisomer appears naturally in proteins. It is not essential to the human diet, since it is synthesized in the body from other metabolites, including glycine. Serine was first obtained from silk protein, a particularly rich source, in 1865 by Emil Cramer. Its name is derived from the Latin for silk, ''sericum''. Serine's structure was establ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Threonine Metabolism
Threonine (symbol Thr or T) is an amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), a carboxyl group (which is in the deprotonated −COO− form under biological conditions), and a side chain containing a hydroxyl group, making it a polar, uncharged amino acid. It is essential in humans, meaning the body cannot synthesize it: it must be obtained from the diet. Threonine is synthesized from aspartate in bacteria such as ''E. coli''. It is encoded by all the codons starting AC (ACU, ACC, ACA, and ACG). Threonine sidechains are often hydrogen bonded; the most common small motifs formed are based on interactions with serine: ST turns, ST motifs (often at the beginning of alpha helices) and ST staples (usually at the middle of alpha helices). Modifications The threonine residue is susceptible to numerous posttranslational modifications. The hydroxyl side-chain can underg ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cysteine Metabolism
Cysteine metabolism refers to the biological pathways that consume or create cysteine. The pathways of different amino acids and other metabolites interweave and overlap to creating complex systems.cysteine is metabolism creating complex systems Human cysteine metabolism In human cysteine metabolism, L-cysteine is consumed in several ways as shown below. L-Cysteine is also consumed in methionine and glutathione metabolism as well as pantothenate/coenzyme A biosynthesis. L-Cysteine is the product of several processes as well. In addition to the reactions below, L-cysteine is also a product of glycine, serine, and threonine metabolism. See also * D-cysteine desulfhydrase * Sulfur metabolism Sulfur is metabolism, metabolized by all organisms, from bacteria and archaea to plants and animals. Sulfur is redox, reduced or redox, oxidized by organisms in a variety of forms. The chemical element, element is present in proteins, organosulfate ... {{DEFAULTSORT:Cysteine Metabolism ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cofactor (biochemistry)
A cofactor is a non-protein chemical compound or metallic ion that is required for an enzyme's role as a catalyst (a catalyst is a substance that increases the rate of a chemical reaction). Cofactors can be considered "helper molecules" that assist in biochemical transformations. The rates at which these happen are characterized in an area of study called enzyme kinetics. Cofactors typically differ from ligands in that they often derive their function by remaining bound. Cofactors can be divided into two types: inorganic ions and complex organic molecules called coenzymes. Coenzymes are mostly derived from vitamins and other organic essential nutrients in small amounts. (Note that some scientists limit the use of the term "cofactor" for inorganic substances; both types are included here.) Coenzymes are further divided into two types. The first is called a "prosthetic group", which consists of a coenzyme that is tightly (or even covalently) and permanently bound to a protein. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyridoxal Phosphate
Pyridoxal phosphate (PLP, pyridoxal 5'-phosphate, P5P), the active form of vitamin B6, is a coenzyme in a variety of enzymatic reactions. The International Union of Biochemistry and Molecular Biology has catalogued more than 140 PLP-dependent activities, corresponding to ~4% of all classified activities. The versatility of PLP arises from its ability to covalently bind the substrate, and then to act as an electrophilic catalyst, thereby stabilizing different types of carbanionic reaction intermediates. Role as a coenzyme PLP acts as a coenzyme in all transamination reactions, and in certain decarboxylation, deamination, and racemization reactions of amino acids. The aldehyde group of PLP forms a Schiff-base linkage (internal aldimine) with the ε-amino group of a specific lysine group of the aminotransferase enzyme. The α-amino group of the amino acid substrate displaces the ε-amino group of the active-site lysine residue in a process known as transaldimination. The resulting ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tertiary Structure
Protein tertiary structure is the three dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains may interact and bond in a number of ways. The interactions and bonds of side chains within a particular protein determine its tertiary structure. The protein tertiary structure is defined by its atomic coordinates. These coordinates may refer either to a protein domain or to the entire tertiary structure.Branden C. and Tooze J. "Introduction to Protein Structure" Garland Publishing, New York. 1990 and 1991. A number of tertiary structures may fold into a quaternary structure.Kyte, J. "Structure in Protein Chemistry." Garland Publishing, New York. 1995. History The science of the tertiary structure of proteins has progressed from one of hypothesis to one of detailed definition. Although Emil Fischer had suggested proteins were made of polypept ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules, such as proteins and nucleic acids. The data, typically obtained by X-ray crystallography, NMR spectroscopy, or, increasingly, cryo-electron microscopy, and submitted by biologists and biochemists from around the world, are freely accessible on the Internet via the websites of its member organisations (PDBe, PDBj, RCSB, and BMRB). The PDB is overseen by an organization called the Worldwide Protein Data Bank, wwPDB. The PDB is a key in areas of structural biology, such as structural genomics. Most major scientific journals and some funding agencies now require scientists to submit their structure data to the PDB. Many other databases use protein structures deposited in the PDB. For example, SCOP and CATH classify protein structures, while PDBsum provides a graphic overview of PDB entries using information from other sources, such as Gene ontology. History Two force ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |