|
Haplotype Estimation
In genetics, haplotype estimation (also known as "phasing") refers to the process of statistical estimation of haplotypes from genotype data. The most common situation arises when genotypes are collected at a set of polymorphic sites from a group of individuals. For example in human genetics, genome-wide association studies collect genotypes in thousands of individuals at between 200,000-5,000,000 SNPs using microarrays. Haplotype estimation methods are used in the analysis of these datasets and allow genotype imputation of alleles from reference databases such as the HapMap Project and the 1000 Genomes Project. Genotypes and haplotypes Genotypes measure the unordered combination of alleles at each site, whereas haplotypes are the two sequences of alleles that have been inherited together from the individual's parents. When there are N heterozygous genotypes present in an individual's set of genotypes, there will be 2^ possible pairs of haplotypes that could underlie the genotypes. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetics
Genetics is the study of genes, genetic variation, and heredity in organisms.Hartl D, Jones E (2005) It is an important branch in biology because heredity is vital to organisms' evolution. Gregor Mendel, a Moravian Augustinian friar working in the 19th century in Brno, was the first to study genetics scientifically. Mendel studied "trait inheritance", patterns in the way traits are handed down from parents to offspring over time. He observed that organisms (pea plants) inherit traits by way of discrete "units of inheritance". This term, still used today, is a somewhat ambiguous definition of what is referred to as a gene. Trait inheritance and molecular inheritance mechanisms of genes are still primary principles of genetics in the 21st century, but modern genetics has expanded to study the function and behavior of genes. Gene structure and function, variation, and distribution are studied within the context of the cell, the organism (e.g. dominance), and within the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
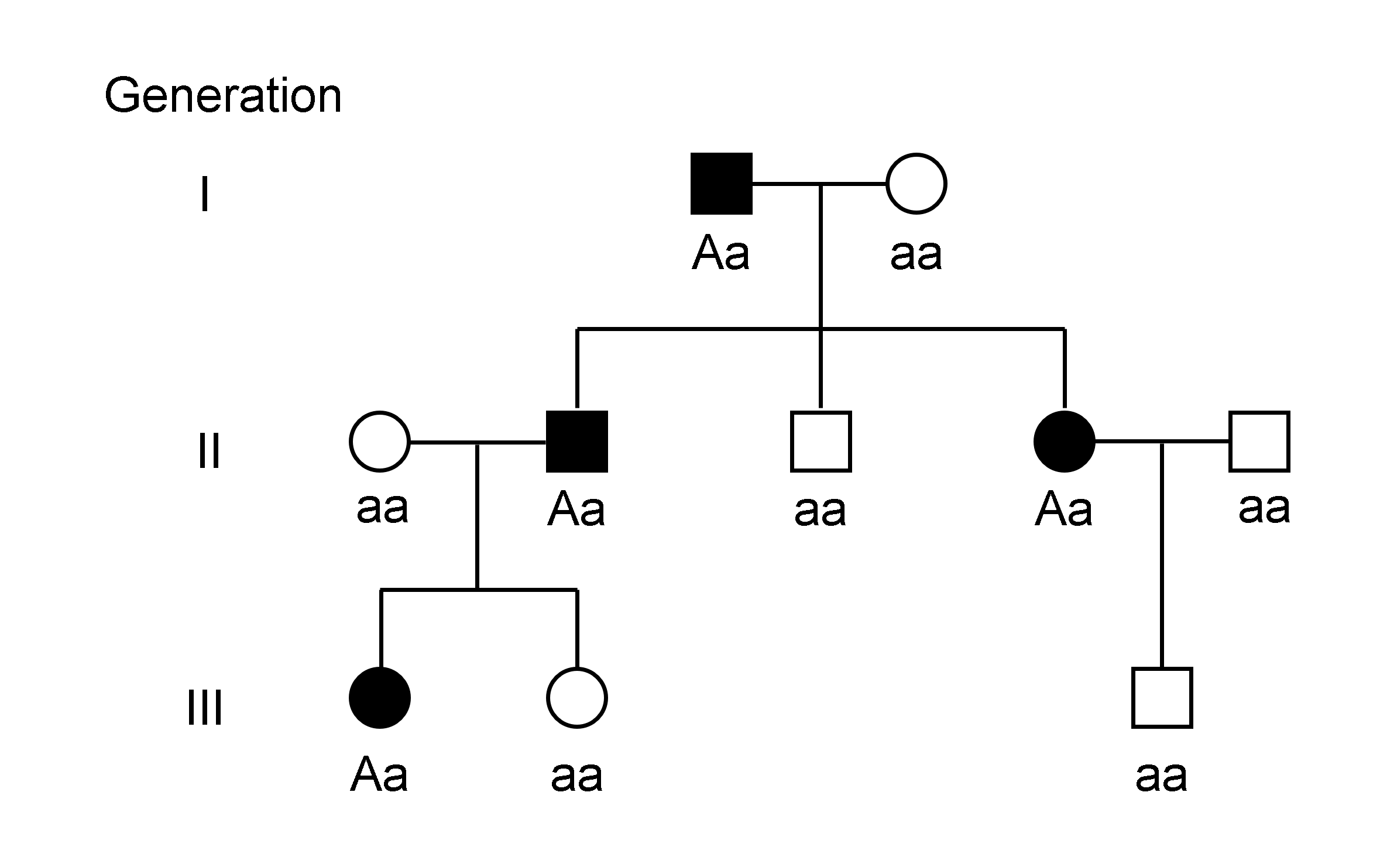
Haplotype
A haplotype ( haploid genotype) is a group of alleles in an organism that are inherited together from a single parent. Many organisms contain genetic material ( DNA) which is inherited from two parents. Normally these organisms have their DNA organized in two sets of pairwise similar chromosomes. The offspring gets one chromosome in each pair from each parent. A set of pairs of chromosomes is called diploid and a set of only one half of each pair is called haploid. The haploid genotype (haplotype) is a genotype that considers the singular chromosomes rather than the pairs of chromosomes. It can be all the chromosomes from one of the parents or a minor part of a chromosome, for example a sequence of 9000 base pairs. However, there are other uses of this term. First, it is used to mean a collection of specific alleles (that is, specific DNA sequences) in a cluster of tightly linked genes on a chromosome that are likely to be inherited together—that is, they are likely to be con ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genotype
The genotype of an organism is its complete set of genetic material. Genotype can also be used to refer to the alleles or variants an individual carries in a particular gene or genetic location. The number of alleles an individual can have in a specific gene depends on the number of copies of each chromosome found in that species, also referred to as ploidy. In diploid species like humans, two full sets of chromosomes are present, meaning each individual has two alleles for any given gene. If both alleles are the same, the genotype is referred to as homozygous. If the alleles are different, the genotype is referred to as heterozygous. Genotype contributes to phenotype, the observable traits and characteristics in an individual or organism. The degree to which genotype affects phenotype depends on the trait. For example, the petal color in a pea plant is exclusively determined by genotype. The petals can be purple or white depending on the alleles present in the pea plant. Howev ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
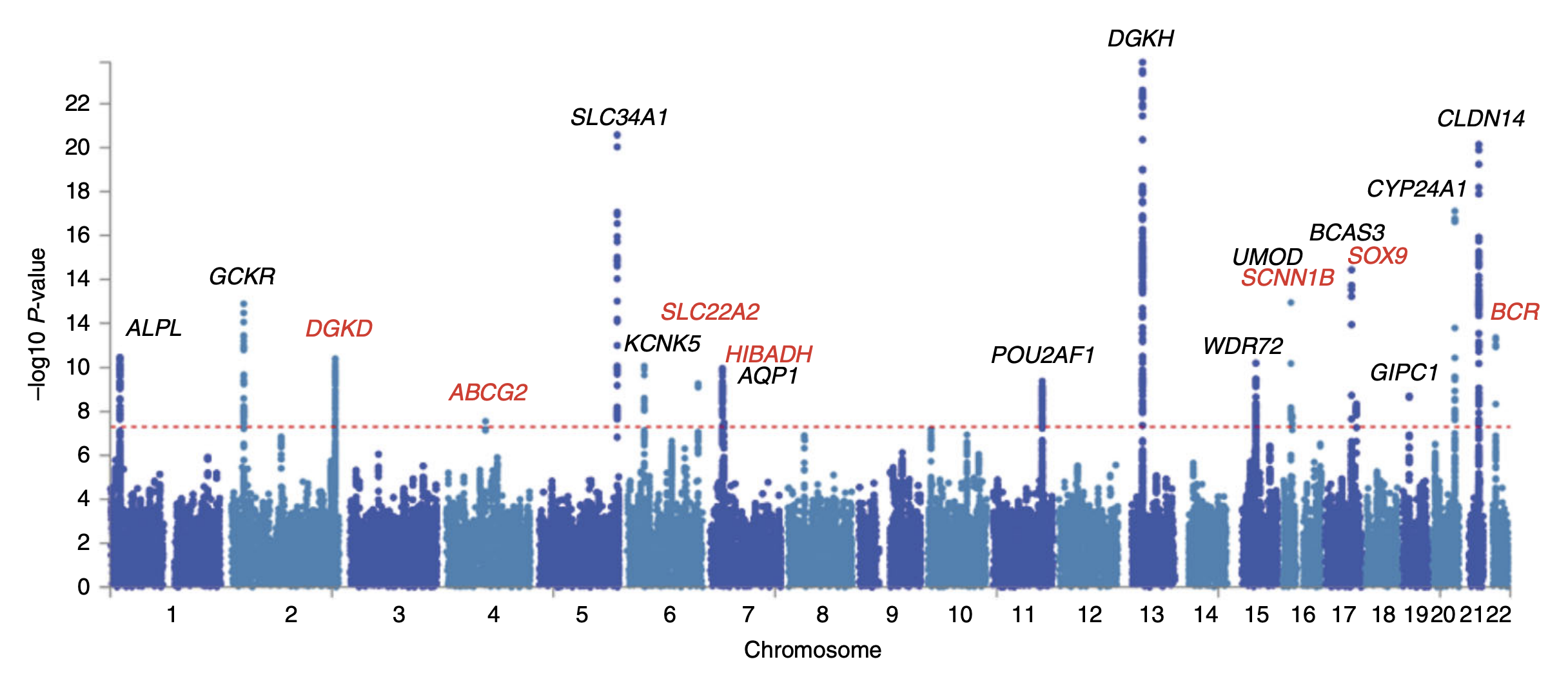
Genome-wide Association Studies
In genomics, a genome-wide association study (GWA study, or GWAS), also known as whole genome association study (WGA study, or WGAS), is an observational study of a genome-wide set of genetic variants in different individuals to see if any variant is associated with a trait. GWA studies typically focus on associations between single-nucleotide polymorphisms (SNPs) and traits like major human diseases, but can equally be applied to any other genetic variants and any other organisms. When applied to human data, GWA studies compare the DNA of participants having varying phenotypes for a particular trait or disease. These participants may be people with a disease (cases) and similar people without the disease (controls), or they may be people with different phenotypes for a particular trait, for example blood pressure. This approach is known as phenotype-first, in which the participants are classified first by their clinical manifestation(s), as opposed to genotype-first. Each per ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Imputation (genetics)
Imputation in genetics refers to the statistical inference of unobserved genotypes. It is achieved by using known haplotypes in a population, for instance from the HapMap or the 1000 Genomes Project in humans, thereby allowing to test for association between a trait of interest (e.g. a disease) and experimentally untyped genetic variants, but whose genotypes have been statistically inferred ("imputed"). Genotype imputation is usually performed on SNPs, the most common kind of genetic variation. Genotype imputation hence helps tremendously in narrowing down the location of probably causal variants in genome-wide association studies, because it increases the SNP density (the genome size remains constant, but the number of genetic variants increases) and thus reduces the distance between two adjacent SNPs. Context In genetic epidemiology and quantitative genetics, researchers aim at identifying genomic locations where variation between individuals is associated with variation in t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HapMap Project
The International HapMap Project was an organization that aimed to develop a haplotype map (HapMap) of the human genome, to describe the common patterns of human genetic variation. HapMap is used to find genetic variants affecting health, disease and responses to drugs and environmental factors. The information produced by the project is made freely available for research. The International HapMap Project is a collaboration among researchers at academic centers, non-profit biomedical research groups and private companies in Canada, China (including Hong Kong), Japan, Nigeria, the United Kingdom, and the United States. It officially started with a meeting on October 27 to 29, 2002, and was expected to take about three years. It comprises two phases; the complete data obtained in Phase I were published on 27 October 2005. The analysis of the Phase II dataset was published in October 2007. The Phase III dataset was released in spring 2009 and the publication presenting the final resul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
The 1000 Genomes Project
The 1000 Genomes Project (abbreviated as 1KGP), launched in January 2008, was an international research effort to establish by far the most detailed catalogue of human genetic variation. Scientists planned to sequence the genomes of at least one thousand anonymous participants from a number of different ethnic groups within the following three years, using newly developed technologies which were faster and less expensive. In 2010, the project finished its pilot phase, which was described in detail in a publication in the journal ''Nature''. In 2012, the sequencing of 1092 genomes was announced in a ''Nature'' publication. In 2015, two papers in ''Nature'' reported results and the completion of the project and opportunities for future research. Many rare variations, restricted to closely related groups, were identified, and eight structural-variation classes were analyzed. The project unites multidisciplinary research teams from institutes around the world, including China, Ita ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heterozygous
Zygosity (the noun, zygote, is from the Greek "yoked," from "yoke") () is the degree to which both copies of a chromosome or gene have the same genetic sequence. In other words, it is the degree of similarity of the alleles in an organism. Most eukaryotes have two matching sets of chromosomes; that is, they are diploid. Diploid organisms have the same loci on each of their two sets of homologous chromosomes except that the sequences at these loci may differ between the two chromosomes in a matching pair and that a few chromosomes may be mismatched as part of a chromosomal sex-determination system. If both alleles of a diploid organism are the same, the organism is homozygous at that locus. If they are different, the organism is heterozygous at that locus. If one allele is missing, it is hemizygous, and, if both alleles are missing, it is nullizygous. The DNA sequence of a gene often varies from one individual to another. These gene variants are called alleles. While some gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EM Algorithm
EM, Em or em may refer to: Arts and entertainment Music * EM, the E major musical scale * Em, the E minor musical scale * Electronic music, music that employs electronic musical instruments and electronic music technology in its production * Encyclopedia Metallum, an online metal music database * Eminem, American rapper Other uses in arts and entertainment * ''Em'' (comic strip), a comic strip by Maria Smedstad Companies and organizations * European Movement, an international lobbying association * Aero Benin (IATA code), a defunct airline * Empire Airlines (IATA code), a charter and cargo airline based in Idaho, US * Erasmus Mundus, an international student-exchange program * ExxonMobil, a large oil company formed from the merger of Exxon and Mobil in 1999 * La République En Marche! (sometimes shortened to "En Marche!"), a major French political party Economics * Emerging markets, nations undergoing rapid industrialization Language and typography Language * M, a letter of t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hidden Markov Model
A hidden Markov model (HMM) is a statistical Markov model in which the system being modeled is assumed to be a Markov process — call it X — with unobservable ("''hidden''") states. As part of the definition, HMM requires that there be an observable process Y whose outcomes are "influenced" by the outcomes of X in a known way. Since X cannot be observed directly, the goal is to learn about X by observing Y. HMM has an additional requirement that the outcome of Y at time t=t_0 must be "influenced" exclusively by the outcome of X at t=t_0 and that the outcomes of X and Y at t handwriting recognition, handwriting, gesture recognition, part-of-speech tagging, musical score following, partial discharges and bioinformatics. Definition Let X_n and Y_n be discrete-time stochastic processes and n\geq 1. The pair (X_n,Y_n) is a ''hidden Markov model'' if * X_n is a Markov process whose behavior is not directly observable ("hidden"); * \operatorname\bigl(Y_n \i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coalescent Theory
Coalescent theory is a model of how alleles sampled from a population may have originated from a common ancestor. In the simplest case, coalescent theory assumes no recombination, no natural selection, and no gene flow or population structure, meaning that each variant is equally likely to have been passed from one generation to the next. The model looks backward in time, merging alleles into a single ancestral copy according to a random process in coalescence events. Under this model, the expected time between successive coalescence events increases almost exponentially back in time (with wide variance). Variance in the model comes from both the random passing of alleles from one generation to the next, and the random occurrence of mutations in these alleles. The mathematical theory of the coalescent was developed independently by several groups in the early 1980s as a natural extension of classical population genetics theory and models, but can be primarily attributed to John K ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
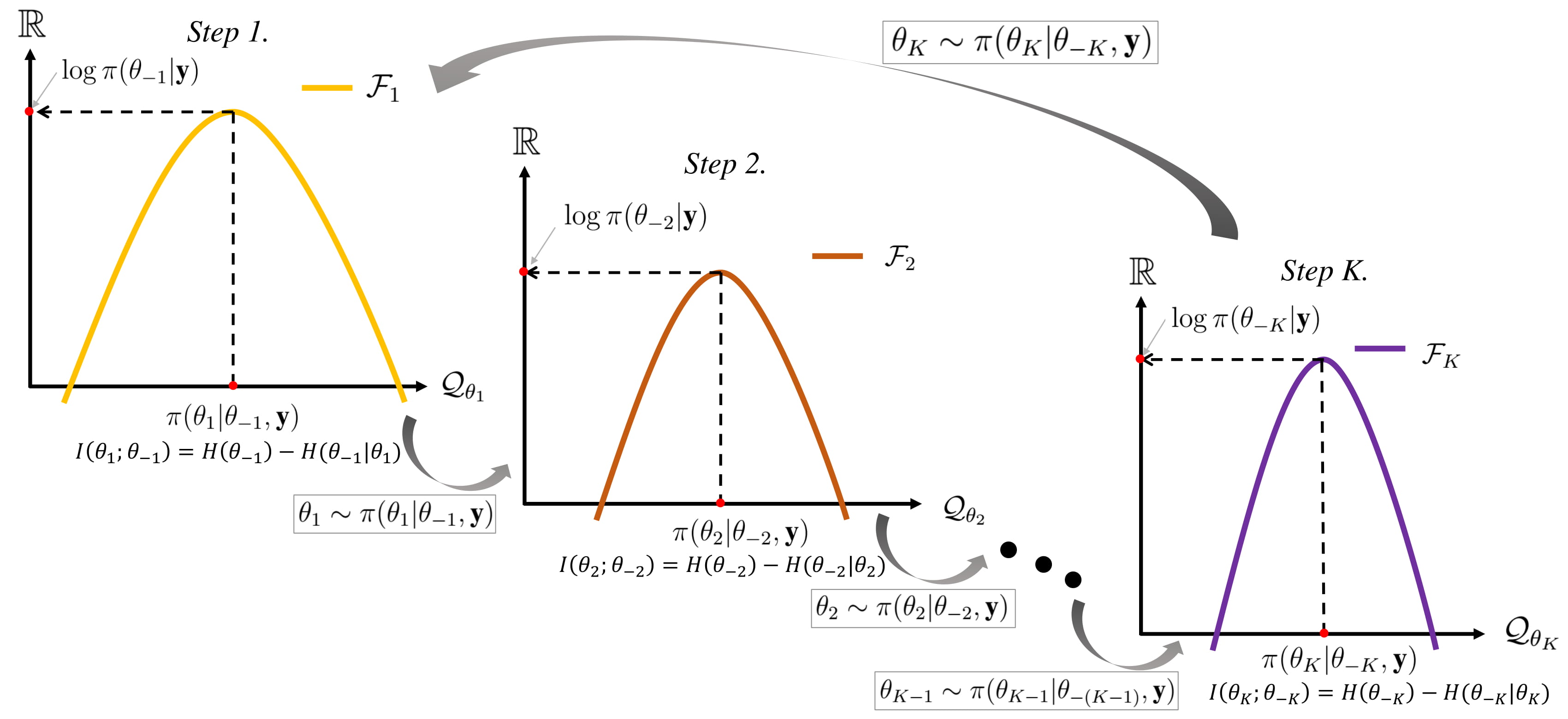
Gibbs Sampling
In statistics, Gibbs sampling or a Gibbs sampler is a Markov chain Monte Carlo (MCMC) algorithm for obtaining a sequence of observations which are approximated from a specified multivariate probability distribution, when direct sampling is difficult. This sequence can be used to approximate the joint distribution (e.g., to generate a histogram of the distribution); to approximate the marginal distribution of one of the variables, or some subset of the variables (for example, the unknown parameters or latent variables); or to compute an integral (such as the expected value of one of the variables). Typically, some of the variables correspond to observations whose values are known, and hence do not need to be sampled. Gibbs sampling is commonly used as a means of statistical inference, especially Bayesian inference. It is a randomized algorithm (i.e. an algorithm that makes use of random numbers), and is an alternative to deterministic algorithms for statistical inferenc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |