|
HMBC
Two-dimensional nuclear magnetic resonance spectroscopy (2D NMR) is a set of nuclear magnetic resonance spectroscopy (NMR) methods which give data plotted in a space defined by two frequency axes rather than one. Types of 2D NMR include correlation spectroscopy (COSY), J-spectroscopy, exchange spectroscopy (EXSY), and nuclear Overhauser effect spectroscopy (NOESY). Two-dimensional NMR spectra provide more information about a molecule than one-dimensional NMR spectra and are especially useful in determining the structure of a molecule, particularly for molecules that are too complicated to work with using one-dimensional NMR. The first two-dimensional experiment, COSY, was proposed by Jean Jeener, a professor at the Université Libre de Bruxelles, in 1971. This experiment was later implemented by Walter P. Aue, Enrico Bartholdi and Richard R. Ernst, who published their work in 1976. Fundamental concepts Each experiment consists of a sequence of radio frequency (RF) pulses with d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Magnetic Resonance Spectroscopy
Nuclear magnetic resonance spectroscopy, most commonly known as NMR spectroscopy or magnetic resonance spectroscopy (MRS), is a spectroscopic technique to observe local magnetic fields around atomic nuclei. The sample is placed in a magnetic field and the NMR signal is produced by excitation of the nuclei sample with radio waves into nuclear magnetic resonance, which is detected with sensitive radio receivers. The intramolecular magnetic field around an atom in a molecule changes the resonance frequency, thus giving access to details of the electronic structure of a molecule and its individual functional groups. As the fields are unique or highly characteristic to individual compounds, in modern organic chemistry practice, NMR spectroscopy is the definitive method to identify monomolecular organic compounds. The principle of NMR usually involves three sequential steps: # The alignment (polarization) of the magnetic nuclear spins in an applied, constant magnetic field B0. # The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HNCOCA Experiment
HNCOCA is a 3D triple-resonance NMR experiment commonly used in the field of protein NMR. The name derives from the experiment's magnetization transfer pathway: The magnetization of the amide proton of an amino acid residue is transferred to the amide nitrogen, and then to the alpha carbon of the previous residue in the protein's amino acid sequence. In contrast, the complementary HNCA experiment transfers magnetization to the alpha carbons of both the starting residue and the previous residue in the sequence. The HNCOCA experiment is used, often in tandem with HNCA, to assign alpha carbon resonance signals to specific residues in the protein. This experiment requires a purified sample of protein prepared with 13C and 15N isotopic labelling, at a concentration greater than 0.1 mM, and is thus generally only applied to recombinant proteins Recombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods of genetic recombination (such as molecular cloning) tha ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HNCA Experiment
HNCA is a 3D triple-resonance NMR experiment commonly used in the field of protein NMR. The name derives from the experiment's magnetization transfer pathway: The magnetization of the amide proton of an amino acid residue is transferred to the amide nitrogen, and then to the alpha carbons of both the starting residue and the previous residue in the protein's amino acid sequence. In contrast, the complementary HNCOCA experiment transfers magnetization only to the alpha carbon of the previous residue. The HNCA experiment is used, often in tandem with HNCOCA, to assign alpha carbon resonance signals to specific residues in the protein. This experiment requires a purified sample of protein prepared with 13C and 15N isotopic labelling, at a concentration greater than 0.1 mM, and is thus generally only applied to recombinant proteins Recombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods of genetic recombination (such as molecular cloning) that bring tog ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Triple Resonance Experiments
Triple resonance experiments are a set of multi-dimensional nuclear magnetic resonance spectroscopy (NMR) experiments that link three types of atomic nuclei, most typically consisting of 1H, 15N and 13C. These experiments are often used to assign specific resonance signals to specific atoms in an isotopically-enriched protein. The technique was first described in papers by Ad Bax, Mitsuhiko Ikura and Lewis Kay in 1990, and further experiments were then added to the suite of experiments. Many of these experiments have since become the standard set of experiments used for sequential assignment of NMR resonances in the determination of protein structure by NMR. They are now an integral part of solution NMR study of proteins, and they may also be used in solid-state NMR. Background There are two main methods of determining protein structure on the atomic level. The first of these is by X-ray crystallography, starting in 1958 when the crystal structure of myoglobin was determined ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dalton (unit)
The dalton or unified atomic mass unit (symbols: Da or u) is a non-SI unit of mass widely used in physics and chemistry. It is defined as of the mass of an unbound neutral atom of carbon-12 in its nuclear and electronic ground state and at rest. The atomic mass constant, denoted ''m''u, is defined identically, giving . This unit is commonly used in physics and chemistry to express the mass of atomic-scale objects, such as atoms, molecules, and elementary particles, both for discrete instances and multiple types of ensemble averages. For example, an atom of helium-4 has a mass of . This is an intrinsic property of the isotope and all helium-4 atoms have the same mass. Acetylsalicylic acid (aspirin), , has an average mass of approximately . However, there are no acetylsalicylic acid molecules with this mass. The two most common masses of individual acetylsalicylic acid molecules are , having the most common isotopes, and , in which one carbon is carbon-13. The molecular ma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Weight
A molecule is a group of two or more atoms held together by attractive forces known as chemical bonds; depending on context, the term may or may not include ions which satisfy this criterion. In quantum physics, organic chemistry, and biochemistry, the distinction from ions is dropped and ''molecule'' is often used when referring to polyatomic ions. A molecule may be homonuclear, that is, it consists of atoms of one chemical element, e.g. two atoms in the oxygen molecule (O2); or it may be heteronuclear, a chemical compound composed of more than one element, e.g. water (two hydrogen atoms and one oxygen atom; H2O). In the kinetic theory of gases, the term ''molecule'' is often used for any gaseous particle regardless of its composition. This relaxes the requirement that a molecule contains two or more atoms, since the noble gases are individual atoms. Atoms and complexes connected by non-covalent interactions, such as hydrogen bonds or ionic bonds, are typically not consi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rotational Correlation Time
Rotational correlation time (\tau_c) is the average time it takes for a molecule to rotate one radian. In solution, rotational correlation times are in the order of picoseconds. For example, the \tau_c = 1.7 ps for water, and 100 ps for a pyrroline nitroxyl radical in a DMSO-water mixture. Rotational correlation times are employed in the measurement of microviscosity (viscosity at the molecular level) and in protein characterization. Rotational correlation times may be measured by rotational (microwave), dielectric, and nuclear magnetic resonance (NMR) spectroscopy. Rotational correlation times of probe molecules in media have been measured by fluorescence lifetime or for radicals, from the linewidths of electron spin resonance Electron paramagnetic resonance (EPR) or electron spin resonance (ESR) spectroscopy is a method for studying materials that have unpaired electrons. The basic concepts of EPR are analogous to those of nuclear magnetic resonance (NMR), but the sp ...s. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequential Walking
Sequential walking is a technique that can be used to solve various 2D NMR spectra. In a 2D experiment, cross peaks must be correlated to the correct nuclei. Using sequential walking, the correct nuclei can be assigned to their crosspeaks. The assigned crosspeaks can give valuable information such as spatial interactions between nuclei. In a NOESY of DNA, for example, each nucleotide has a different chemical shift associated with it. In general, A's are more downstream, T's are more upstream, and C's and G's are intermediate. Each nucleotide has protons on the deoxyribose sugar, which can be assigned using sequential walking. To do this, the first nucleotide in the sequence must be detected. Knowing the DNA sequence helps, but in general the first nucleotide Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein NMR
Nuclear magnetic resonance spectroscopy of proteins (usually abbreviated protein NMR) is a field of structural biology in which NMR spectroscopy is used to obtain information about the structure and dynamics of proteins, and also nucleic acids, and their complexes. The field was pioneered by Richard R. Ernst and Kurt Wüthrich at the ETH, and by Ad Bax, Marius Clore, Angela Gronenborn at the NIH, and Gerhard Wagner at Harvard University, among others. Structure determination by NMR spectroscopy usually consists of several phases, each using a separate set of highly specialized techniques. The sample is prepared, measurements are made, interpretive approaches are applied, and a structure is calculated and validated. NMR involves the quantum-mechanical properties of the central core ("nucleus") of the atom. These properties depend on the local molecular environment, and their measurement provides a map of how the atoms are linked chemically, how close they are in space, and how ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Spin–lattice Relaxation
During nuclear magnetic resonance observations, spin–lattice relaxation is the mechanism by which the longitudinal component of the total nuclear magnetic moment vector (parallel to the constant magnetic field) exponentially relaxes from a higher energy, non-equilibrium state to thermodynamic equilibrium with its surroundings (the "lattice"). It is characterized by the spin–lattice relaxation time, a time constant known as ''T1''. There is a different parameter, ''T2'', the spin-spin relaxation time, which concerns the exponential relaxation of the transverse component of the nuclear magnetization vector ( to the external magnetic field). Measuring the variation of ''T1'' and ''T2'' in different materials is the basis for some magnetic resonance imaging techniques. Nuclear physics ''T1'' relaxation or longitudinal relaxation curve ''T1'' characterizes the rate at which the longitudinal ''Mz'' component of the magnetization vector recovers exponentially towards its thermodyna ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
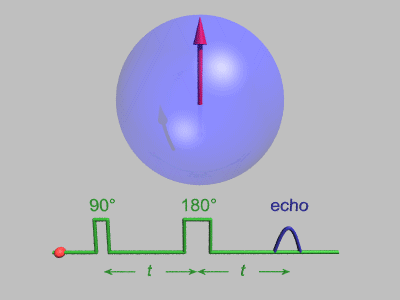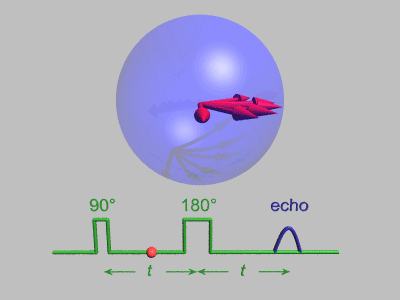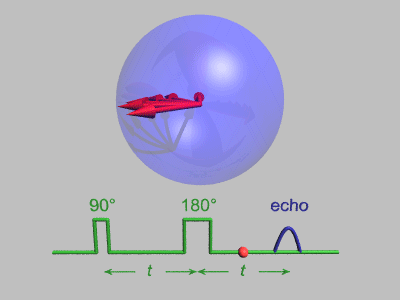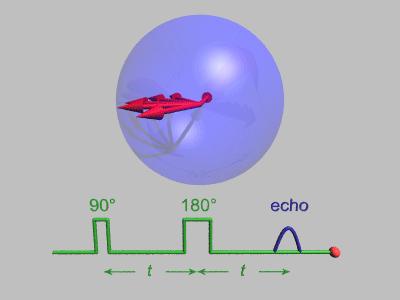
Spin Echo
In magnetic resonance, a spin echo or Hahn echo is the refocusing of spin magnetisation by a pulse of resonant electromagnetic radiation. Modern nuclear magnetic resonance (NMR) and magnetic resonance imaging (MRI) make use of this effect. The NMR signal observed following an initial excitation pulse decays with time due to both spin relaxation and any ''inhomogeneous'' effects which cause spins in the sample to precess at different rates. The first of these, relaxation, leads to an irreversible loss of magnetisation. But the inhomogeneous dephasing can be removed by applying a 180° ''inversion'' pulse that inverts the magnetisation vectors. Examples of inhomogeneous effects include a magnetic field gradient and a distribution of chemical shifts. If the inversion pulse is applied after a period ''t'' of dephasing, the inhomogeneous evolution will rephase to form an echo at time 2''t''. In simple cases, the intensity of the echo relative to the initial signal is given by ''e– ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |