|
High Potential Iron–sulfur Protein
High potential iron-sulfur proteins (HIPIP) are a specific class of high-redox potential 4Fe-4S ferredoxins that functions in anaerobic electron transport and which occurs in photosynthetic bacteria and in '' Paracoccus denitrificans''. The HiPIPs are small proteins which show significant variation in their sequences, their sizes (from 63 to 85 amino acids), and in their oxidation- reduction potentials. As shown in the following schematic representation the iron-sulfur cluster is bound by four conserved cysteine residues. 4Fe-4S cluster , , , , xxxxxxxxxxxxxxxxxxxCxCxxxxxxxCxxxxxCxxxx C: conserved cysteine involved in the binding of the iron-sulfur cluster. e4S4clusters The e4S4clusters are abundant cofactors of metalloproteins. They participate in electron-transfer sequences. The core structure for the e4S4cluster is a cube with alternating Fe and S vertices. These clusters exist in two oxidation st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ferredoxin
Ferredoxins (from Latin ''ferrum'': iron + redox, often abbreviated "fd") are iron–sulfur proteins that mediate electron transfer in a range of metabolic reactions. The term "ferredoxin" was coined by D.C. Wharton of the DuPont Co. and applied to the "iron protein" first purified in 1962 by Mortenson, Valentine, and Carnahan from the anaerobic bacterium '' Clostridium pasteurianum''. Another redox protein, isolated from spinach chloroplasts, was termed "chloroplast ferredoxin". The chloroplast ferredoxin is involved in both cyclic and non-cyclic photophosphorylation reactions of photosynthesis. In non-cyclic photophosphorylation, ferredoxin is the last electron acceptor thus reducing the enzyme NADP+ reductase. It accepts electrons produced from sunlight- excited chlorophyll and transfers them to the enzyme ferredoxin: NADP+ oxidoreductase . Ferredoxins are small proteins containing iron and sulfur atoms organized as iron–sulfur clusters. These biological " capacitors" can ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Paracoccus Denitrificans
''Paracoccus denitrificans'', is a coccoid bacterium known for its nitrate reducing properties, its ability to replicate under conditions of hypergravity and for being a relative of the eukaryotic mitochondrion (endosymbiotic theory). Description ''Paracoccus denitrificans'', is a gram-negative, coccus, non-motile, denitrifying (nitrate-reducing) bacterium. It is typically a rod-shaped bacterium but assumes spherical shapes during the stationary phase. Like all gram-negative bacteria, it has a double membrane with a cell wall. Formerly known as ''Micrococcus denitrificans'', it was first isolated in 1910 by Martinus Beijerinck, a Dutch microbiologist. The bacterium was reclassified in 1969 to ''Paracoccus denitrificans'' by D.H. Davis. The genome of ''P. denitrificans'' was sequenced in 2004. Ecology and ecological applications Metabolically ''Paracoccus denitrificans'' is very flexible and has been recorded in soil in both aerobic or anaerobic environments. The microbe also ha ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromatium
''Chromatium'' is a genus of photoautotrophic Gram-negative bacteria which are found in water. The cells are straight rod-shaped or slightly curved. They belong to the purple sulfur bacteria and oxidize sulfide to produce sulfur which is deposited in intracellular granules of the cytoplasm In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. The ....George M. Garrity: '' Bergey's Manual of Systematic Bacteriology''. 2. Auflage. Springer, New York, 2005, Volume 2: ''The Proteobacteria, Part B: The Gammaproteobacteria'' References External links''Chromatium''J.P. Euzéby: List of Prokaryotic names with Standing in Nomenclature Chromatiales Phototrophic bacteria Bacteria genera {{Chromatiales-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
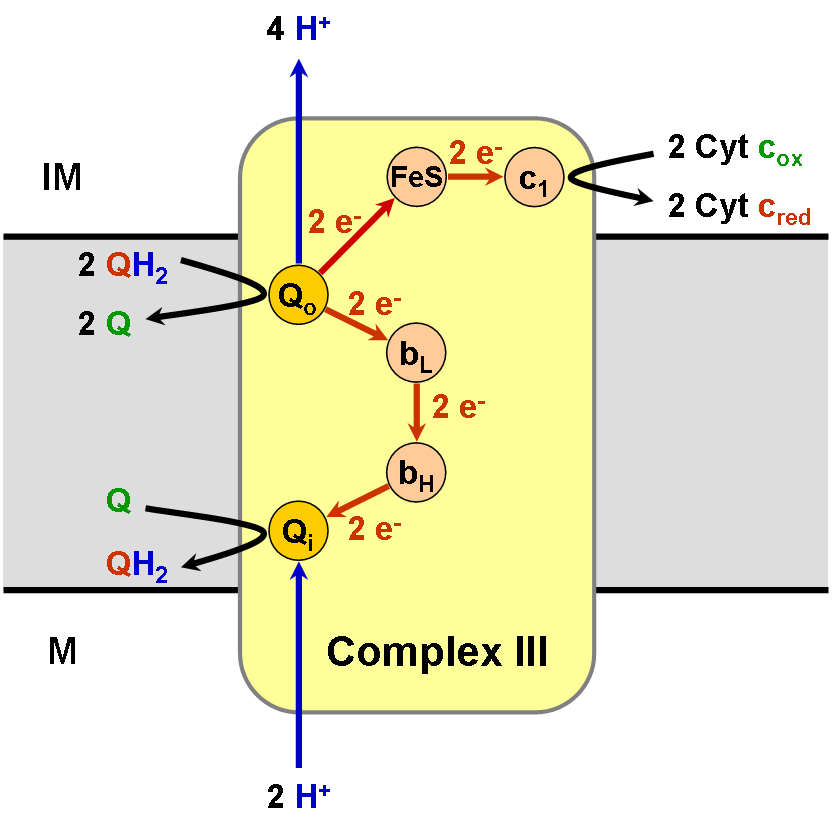
Coenzyme Q – Cytochrome C Reductase
The coenzyme Q : cytochrome ''c'' – oxidoreductase, sometimes called the cytochrome ''bc''1 complex, and at other times complex III, is the third complex in the electron transport chain (), playing a critical role in biochemical generation of ATP (oxidative phosphorylation). Complex III is a multisubunit transmembrane protein encoded by both the mitochondrial (cytochrome b) and the nuclear genomes (all other subunits). Complex III is present in the mitochondria of all animals and all aerobic eukaryotes and the inner membranes of most eubacteria. Mutations in Complex III cause exercise intolerance as well as multisystem disorders. The bc1 complex contains 11 subunits, 3 respiratory subunits (cytochrome B, cytochrome C1, Rieske protein), 2 core proteins and 6 low-molecular weight proteins. Ubiquinol—cytochrome-c reductase catalyzes the chemical reaction :QH2 + 2 ferricytochrome c \rightleftharpoons Q + 2 ferrocytochrome c + 2 H+ Thus, the two substrates of this enzyme ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PROSITE
PROSITE is a protein database. It consists of entries describing the protein families, domains and functional sites as well as amino acid patterns and profiles in them. These are manually curated by a team of the Swiss Institute of Bioinformatics and tightly integrated into Swiss-Prot protein annotation. PROSITE was created in 1988 by Amos Bairoch, who directed the group for more than 20 years. Since July 2018, the director of PROSITE and Swiss-Prot is Alan Bridge. PROSITE's uses include identifying possible functions of newly discovered proteins and analysis of known proteins for previously undetermined activity. Properties from well-studied genes can be propagated to biologically related organisms, and for different or poorly known genes biochemical functions can be predicted from similarities. PROSITE offers tools for protein sequence analysis and motif detection (see sequence motif, PROSITE patterns). It is part of the ExPASy proteomics analysis servers. The database ProR ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Families
A protein family is a group of evolutionarily related proteins. In many cases, a protein family has a corresponding gene family, in which each gene encodes a corresponding protein with a 1:1 relationship. The term "protein family" should not be confused with family as it is used in taxonomy. Proteins in a family descend from a common ancestor and typically have similar three-dimensional structures, functions, and significant sequence similarity. The most important of these is sequence similarity (usually amino-acid sequence), since it is the strictest indicator of homology and therefore the clearest indicator of common ancestry. A fairly well developed framework exists for evaluating the significance of similarity between a group of sequences using sequence alignment methods. Proteins that do not share a common ancestor are very unlikely to show statistically significant sequence similarity, making sequence alignment a powerful tool for identifying the members of protein familie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |