|
Gap Penalty
A Gap penalty is a method of scoring alignments of two or more sequences. When aligning sequences, introducing gaps in the sequences can allow an alignment algorithm to match more terms than a gap-less alignment can. However, minimizing gaps in an alignment is important to create a useful alignment. Too many gaps can cause an alignment to become meaningless. Gap penalties are used to adjust alignment scores based on the number and length of gaps. The five main types of gap penalties are constant, linear, affine, convex, and profile-based. Applications * Genetic sequence alignment - In bioinformatics, gaps are used to account for genetic mutations occurring from insertions or deletions in the sequence, sometimes referred to as ''indels''. Insertions or deletions can occur due to single mutations, unbalanced crossover in meiosis, slipped strand mispairing, and chromosomal translocation. The notion of a gap in an alignment is important in many biological applications, since the i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
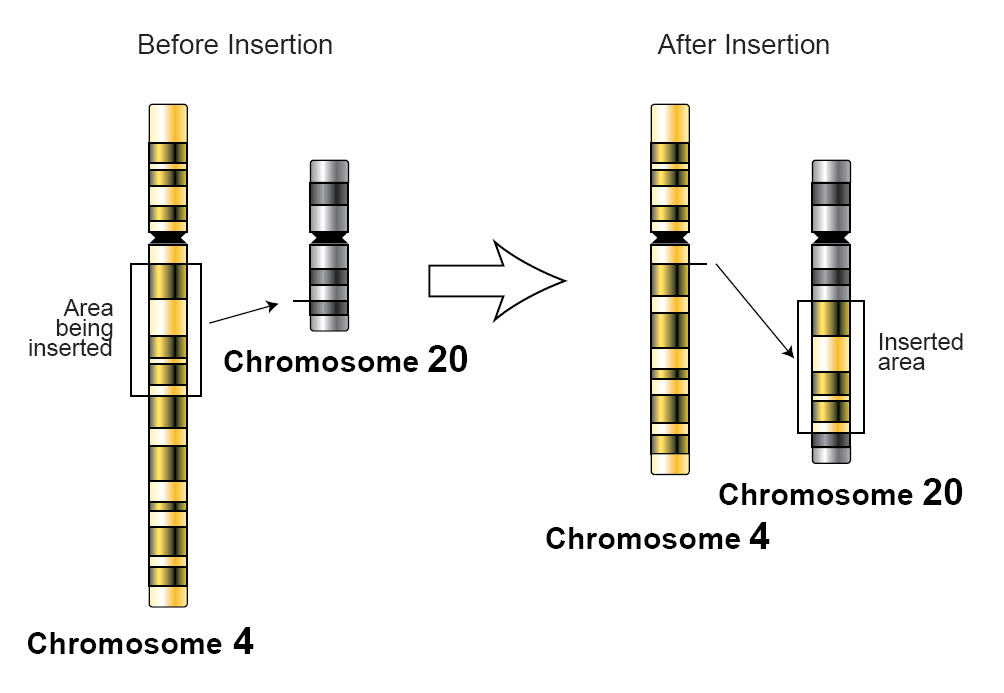
Insertion (genetics)
In genetics, an insertion (also called an insertion mutation) is the addition of one or more nucleotide base pairs into a DNA sequence. This can often happen in microsatellite regions due to the DNA polymerase slipping. Insertions can be anywhere in size from one base pair incorrectly inserted into a DNA sequence to a section of one chromosome inserted into another. The mechanism of the smallest single base insertion mutations is believed to be through base-pair separation between the template and primer strands followed by non-neighbor base stacking, which can occur locally within the DNA polymerase active site. On a chromosome level, an ''insertion'' refers to the insertion of a larger sequence into a chromosome. This can happen due to unequal crossover during meiosis. N region addition is the addition of non-coded nucleotides during recombination by terminal deoxynucleotidyl transferase. P nucleotide insertion is the insertion of palindromic sequences encoded by the ends ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Substitution Matrix
In bioinformatics and evolutionary biology, a substitution matrix describes the frequency at which a character in a nucleotide sequence or a protein sequence changes to other character states over evolutionary time. The information is often in the form of log odds of finding two specific character states aligned and depends on the assumed number of evolutionary changes or sequence dissimilarity between compared sequences. It is an application of a stochastic matrix. Substitution matrices are usually seen in the context of amino acid or DNA sequence alignments, where they are used to calculate similarity scores between the aligned sequences. Background In the process of evolution, from one generation to the next the amino acid sequences of an organism's proteins are gradually altered through the action of DNA mutations. For example, the sequence ALEIRYLRD could mutate into the sequence ALEINYLRD in one step, and possibly AQEINYQRD over a longer period of evolutionar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Comparison Of Gap Penalty Funcitons
Comparison or comparing is the act of evaluating two or more things by determining the relevant, comparable characteristics of each thing, and then determining which characteristics of each are similar to the other, which are different, and to what degree. Where characteristics are different, the differences may then be evaluated to determine which thing is best suited for a particular purpose. The description of similarities and differences found between the two things is also called a comparison. Comparison can take many distinct forms, varying by field: To compare things, they must have characteristics that are similar enough in relevant ways to merit comparison. If two things are too different to compare in a useful way, an attempt to compare them is colloquially referred to in English as "comparing apples and oranges." Comparison is widely used in society, in science and in the arts. General usage Comparison is a natural activity, which even animals engage in when deci ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cancer
Cancer is a group of diseases involving abnormal cell growth with the potential to invade or spread to other parts of the body. These contrast with benign tumors, which do not spread. Possible signs and symptoms include a lump, abnormal bleeding, prolonged cough, unexplained weight loss, and a change in bowel movements. While these symptoms may indicate cancer, they can also have other causes. Over 100 types of cancers affect humans. Tobacco use is the cause of about 22% of cancer deaths. Another 10% are due to obesity, poor diet, lack of physical activity or excessive drinking of alcohol. Other factors include certain infections, exposure to ionizing radiation, and environmental pollutants. In the developing world, 15% of cancers are due to infections such as '' Helicobacter pylori'', hepatitis B, hepatitis C, human papillomavirus infection, Epstein–Barr virus and human immunodeficiency virus (HIV). These factors act, at least partly, by changing the genes o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
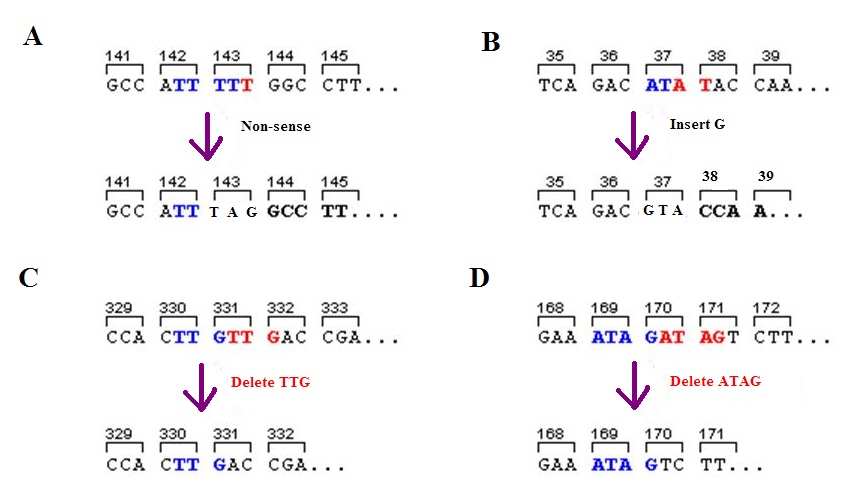
Frameshift Mutation
A frameshift mutation (also called a framing error or a reading frame shift) is a genetic mutation caused by indels (gene insertion, insertions or genetic deletion, deletions) of a number of nucleotides in a DNA sequence that is not divisible by three. Due to the triplet nature of gene expression by codons, the insertion or deletion can change the reading frame (the grouping of the codons), resulting in a completely different Translation (genetics), translation from the original. The earlier in the sequence the deletion or insertion occurs, the more altered the protein. A frameshift mutation is not the same as a single-nucleotide polymorphism in which a nucleotide is replaced, rather than inserted or deleted. A frameshift mutation will in general cause the reading of the codons after the mutation to code for different amino acids. The frameshift mutation will also alter the first stop codon ("UAA", "UGA" or "UAG") encountered in the sequence. The polypeptide being created could be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Indels
Indel is a molecular biology term for an insertion or deletion of bases in the genome of an organism. It is classified among small genetic variations, measuring from 1 to 10 000 base pairs in length, including insertion and deletion events that may be separated by many years, and may not be related to each other in any way. A microindel is defined as an indel that results in a net change of 1 to 50 nucleotides. In coding regions of the genome, unless the length of an indel is a multiple of 3, it will produce a frameshift mutation. For example, a common microindel which results in a frameshift causes Bloom syndrome in the Jewish or Japanese population. Indels can be contrasted with a point mutation. An indel inserts or deletes nucleotides from a sequence, while a point mutation is a form of substitution that ''replaces'' one of the nucleotides without changing the overall number in the DNA. Indels can also be contrasted with Tandem Base Mutations (TBM), which may result from ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential. DNA is made up of a double helix of two complementary strands. The double helix describes the appearance of a double-stranded DNA which is thus composed of two linear strands that run opposite to each other and twist together to form. During replication, these strands are separated. Each strand of the original DNA molecule then serves as a template for the production of its counterpart, a process referred to as semiconservative replication. As a result of semi-conservativ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BLOSUM
In bioinformatics, the BLOSUM (BLOcks SUbstitution Matrix) matrix is a substitution matrix used for sequence alignment of proteins. BLOSUM matrices are used to score alignments between evolutionarily divergent protein sequences. They are based on local alignments. BLOSUM matrices were first introduced in a paper by Steven Henikoff and Jorja Henikoff. They scanned the BLOCKS database for very conserved regions of protein families (that do not have gaps in the sequence alignment) and then counted the relative frequencies of amino acids and their substitution probabilities. Then, they calculated a log-odds score for each of the 210 possible substitution pairs of the 20 standard amino acids. All BLOSUM matrices are based on observed alignments; they are not extrapolated from comparisons of closely related proteins like the PAM Matrices. Biological background The genetic instructions of every replicating cell in a living organism are contained within its DNA. Throughout the cell's ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
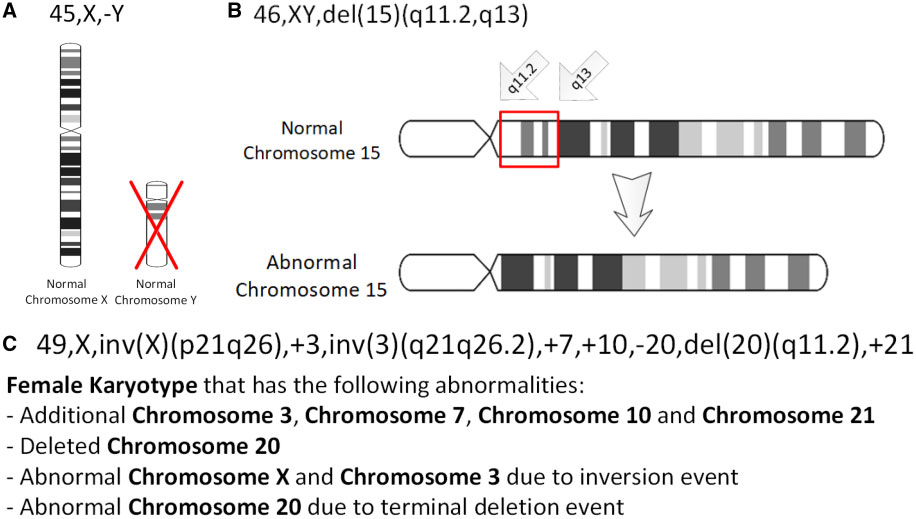
Deletion (genetics)
In genetics, a deletion (also called gene deletion, deficiency, or deletion mutation) (sign: Δ) is a mutation (a genetic aberration) in which a part of a chromosome or a sequence of DNA is left out during DNA replication. Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome. Some chromosomes have fragile spots where breaks occur which result in the deletion of a part of chromosome. The breaks can be induced by heat, viruses, radiations, chemicals. When a chromosome breaks, a part of it is deleted or lost, the missing piece of chromosome is referred to as deletion or a deficiency. For synapsis to occur between a chromosome with a large intercalary deficiency and a normal complete homolog, the unpaired region of the normal homolog must loop out of the linear structure into a deletion or compensation loop. The smallest single base deletion mutations occur by a single base flipping in the template DNA, followed by template DNA strand slip ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Alignment
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residues in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dynamic Programming
Dynamic programming is both a mathematical optimization method and a computer programming method. The method was developed by Richard Bellman in the 1950s and has found applications in numerous fields, from aerospace engineering to economics. In both contexts it refers to simplifying a complicated problem by breaking it down into simpler sub-problems in a recursive manner. While some decision problems cannot be taken apart this way, decisions that span several points in time do often break apart recursively. Likewise, in computer science, if a problem can be solved optimally by breaking it into sub-problems and then recursively finding the optimal solutions to the sub-problems, then it is said to have '' optimal substructure''. If sub-problems can be nested recursively inside larger problems, so that dynamic programming methods are applicable, then there is a relation between the value of the larger problem and the values of the sub-problems.Cormen, T. H.; Leiserson, C. E.; Riv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |