|
Gal4
The Gal4 transcription factor is a positive regulator of gene expression of galactose-induced genes. This protein represents a large fungal family of transcription factors, Gal4 family, which includes over 50 members in the yeast ''Saccharomyces cerevisiae'' e.g. Oaf1, Pip2, Pdr1, Pdr3, Leu3. Gal4 recognizes genes with UAS, an upstream activating sequence, and activates them. In yeast cells, the principal targets are GAL1 (galactokinase), GAL10 (UDP-glucose 4-epimerase), and GAL7 (galactose-1-phosphate uridylyltransferase), three enzymes required for galactose metabolism. This binding has also proven useful in constructing the GAL4/UAS system, a technique for controlling expression in insects. In yeast, Gal4 is by default repressed by Gal80, and activated in the presence of galactose as Gal3 binds away Gal80. Domains Two executive domains, DNA binding and activation domains, provide key function of the Gal4 protein conforming to most of the transcription factors. DNA binding ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gal4 9aaTAD
The Gal4 transcription factor is a positive regulator of gene expression of galactose-induced genes. This protein represents a large fungal family of transcription factors, Gal4 family, which includes over 50 members in the yeast ''Saccharomyces cerevisiae'' e.g. Oaf1, Pip2, Pdr1, Pdr3, Leu3. Gal4 recognizes genes with UAS, an upstream activating sequence, and activates them. In yeast cells, the principal targets are GAL1 (galactokinase), GAL10 (UDP-glucose 4-epimerase), and GAL7 (galactose-1-phosphate uridylyltransferase), three enzymes required for galactose metabolism. This binding has also proven useful in constructing the GAL4/UAS system, a technique for controlling expression in insects. In yeast, Gal4 is by default repressed by Gal80, and activated in the presence of galactose as Gal3 binds away Gal80. Domains Two executive domains, DNA binding and activation domains, provide key function of the Gal4 protein conforming to most of the transcription factors. DNA binding ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
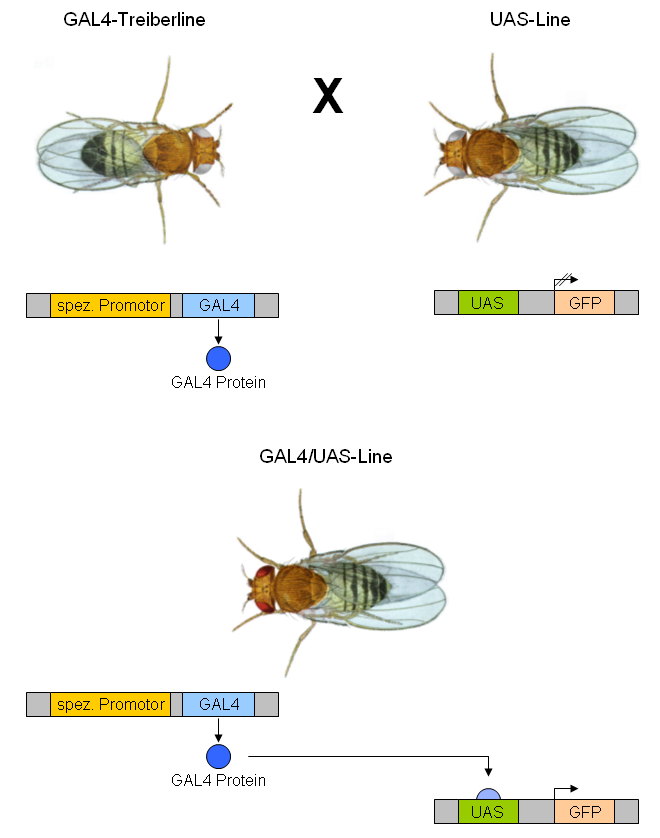
GAL4/UAS System
The GAL4-UAS system is a biochemical method used to study gene expression and function in organisms such as the fruit fly. It is based on the finding by Hitoshi Kakidani and Mark Ptashne, and Nicholas Webster and Pierre Chambon in 1988 that Gal4 binding to UAS sequences activates gene expression. The method was introduced into flies by Andrea Brand and Norbert Perrimon in 1993 and is considered a powerful technique for studying the expression of genes. The system has two parts: the Gal4 gene, encoding the yeast transcription activator protein Gal4, and the UAS (Upstream Activation Sequence), an enhancer to which GAL4 specifically binds to activate gene transcription. Overview The Gal4 system allows separation of the problems of defining which cells express a gene or protein and what the experimenter wants to do with this knowledge. Geneticists have created genetic variants of model organisms (typically fruit flies), called ''GAL4 lines'', each of which expresses GAL4 in some su ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transactivation Domain
The transactivation domain or trans-activating domain (TAD) is a transcription factor scaffold domain which contains binding sites for other proteins such as transcription coregulators. These binding sites are frequently referred to as activation functions (AFs). TADs are named after their amino acid composition. These amino acids are either essential for the activity or simply the most abundant in the TAD. Transactivation by the Gal4 transcription factor is mediated by acidic amino acids, whereas hydrophobic residues in Gcn4 play a similar role. Hence, the TADs in Gal4 and Gcn4 are referred to as acidic or hydrophobic, respectively. In general we can distinguish four classes of TADs: * acidic domains (called also “acid blobs” or “negative noodles", rich in D and E amino acids, present in Gal4, Gcn4 and VP16). * glutamine-rich domains (contains multiple repetitions like "QQQXXXQQQ", present in SP1) * proline-rich domains (contains repetitions like "PPPXXXPPP" present in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MED15
Mediator of RNA polymerase II transcription subunit 15, also known as Gal11, Spt13 in yeast and PCQAP, ARC105, or TIG-1 in humans is a protein encoded by the ''MED15'' gene. Function MED15 is a general transcriptional cofactor of the mediator complex involved in RNA polymerase II dependent transcription, originally called Gal11 and Spt13 and found in yeast as an essential factor for Gal4 dependent transactivation by T.Fukasawa and F.Winston labs. Transcription factors Gcn4, Pho4, Msn2, Ino2, members of the Gal4 family - Gal4, Oaf1, Pdr1, and viral VP16 have been reported to interact with yeast MED15. ; ; ; ; ; Most of these transcription factors share the same transactivation domain, 9aaTAD, which directly interacts with KIX domain of the MED15. ; ; ; ; ; Furthermore, human MED15 cooperates in mediator complex (previously known as PC2, ARC, or DRIP) with transcription factors like VP16 and SREBP. Human SREBP regulates sterol responsive gene expression, and this regulator ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Two-hybrid Screening
Two-hybrid screening (originally known as yeast two-hybrid system or Y2H) is a molecular biology technique used to discover protein–protein interactions (PPIs) and protein–DNA interactions by testing for physical interactions (such as binding) between two proteins or a single protein and a DNA molecule, respectively. The premise behind the test is the activation of downstream reporter gene(s) by the binding of a transcription factor onto an upstream activating sequence (UAS). For two-hybrid screening, the transcription factor is split into two separate fragments, called the DNA-binding domain (DBD or often also abbreviated as BD) and activating domain (AD). The BD is the domain responsible for binding to the UAS and the AD is the domain responsible for the activation of transcription. The Y2H is thus a protein-fragment complementation assay. History Pioneered by Stanley Fields and Ok-Kyu Song in 1989, the technique was originally designed to detect protein–protein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Zinc Finger
A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) in order to stabilize the fold. It was originally coined to describe the finger-like appearance of a hypothesized structure from the African clawed frog (''Xenopus laevis'') transcription factor IIIA. However, it has been found to encompass a wide variety of differing protein structures in eukaryotic cells. ''Xenopus laevis'' TFIIIA was originally demonstrated to contain zinc and require the metal for function in 1983, the first such reported zinc requirement for a gene regulatory protein followed soon thereafter by the Krüppel factor in ''Drosophila''. It often appears as a metal-binding domain in multi-domain proteins. Proteins that contain zinc fingers (zinc finger proteins) are classified into several different structural families. Unlike many other clearly defined supersecondary structures such as Greek keys or β hairpins, there are a number of t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Upstream Activating Sequence
An upstream activating sequence or upstream activation sequence (UAS) is a cis-acting regulatory sequence. It is distinct from the promoter and increases the expression of a neighbouring gene. Due to its essential role in activating transcription, the upstream activating sequence is often considered to be analogous to the function of the enhancer in multicellular eukaryotes. Upstream activation sequences are a crucial part of induction, enhancing the expression of the protein of interest through increased transcriptional activity. The upstream activation sequence is found adjacently upstream to a minimal promoter (TATA box) and serves as a binding site for transactivators. If the transcriptional transactivator does not bind to the UAS in the proper orientation then transcription cannot begin. To further understand the function of an upstream activation sequence, it is beneficial to see its role in the cascade of events that lead to transcription activation. The pathway begins when ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Coregulators
In molecular biology and genetics, transcription coregulators are proteins that interact with transcription factors to either activate or repress the transcription of specific genes. Transcription coregulators that activate gene transcription are referred to as coactivators while those that repress are known as corepressors. The mechanism of action of transcription coregulators is to modify chromatin structure and thereby make the associated DNA more or less accessible to transcription. In humans several dozen to several hundred coregulators are known, depending on the level of confidence with which the characterisation of a protein as a coregulator can be made. One class of transcription coregulators modifies chromatin structure through covalent modification of histones. A second ATP dependent class modifies the conformation of chromatin. Histone acetyltransferases Nuclear DNA is normally tightly wrapped around histones rendering the DNA inaccessible to the general transcr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Leavening Agents
In cooking, a leavening agent () or raising agent, also called a leaven () or leavener, is any one of a number of substances used in doughs and batters that cause a foaming action (gas bubbles) that lightens and softens the mixture. An alternative or supplement to leavening agents is mechanical action by which air is incorporated (i.e. kneading). Leavening agents can be biological or synthetic chemical compounds. The gas produced is often carbon dioxide, or occasionally hydrogen. When a dough or batter is mixed, the starch in the flour and the water in the dough form a matrix (often supported further by proteins like gluten or polysaccharides, such as pentosans or xanthan gum). The starch then gelatinizes and sets, leaving gas bubbles that remain. Biological leavening agents * ''Saccharomyces cerevisiae'' producing carbon dioxide found in: ** baker's yeast ** Beer barm (unpasteurised—live yeast) ** ginger beer ** kefir ** sourdough starter * ''Clostridium perfringens'' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Yeasts Used In Brewing
Yeasts are eukaryotic, single-celled microorganisms classified as members of the fungus kingdom. The first yeast originated hundreds of millions of years ago, and at least 1,500 species are currently recognized. They are estimated to constitute 1% of all described fungal species. Yeasts are unicellular organisms that evolved from multicellular ancestors, with some species having the ability to develop multicellular characteristics by forming strings of connected budding cells known as pseudohyphae or false hyphae. Yeast sizes vary greatly, depending on species and environment, typically measuring 3–4 µm in diameter, although some yeasts can grow to 40 µm in size. Most yeasts reproduce asexually by mitosis, and many do so by the asymmetric division process known as budding. With their single-celled growth habit, yeasts can be contrasted with molds, which grow hyphae. Fungal species that can take both forms (depending on temperature or other conditions) are called ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Osmophiles
An osmophile is a microorganism adapted to environments with high osmotic pressures, such as high sugar concentrations. Osmophiles are similar to halophiles (salt-loving organisms) in that a critical aspect of both types of environment is their low water activity, aW. High sugar concentrations represent a growth-limiting factor for many microorganisms, yet osmophiles protect themselves against this high osmotic pressure by the synthesis of osmoprotectants such as alcohols and amino acids. Many osmophilic microorganisms are yeast Yeasts are eukaryotic, single-celled microorganisms classified as members of the fungus kingdom. The first yeast originated hundreds of millions of years ago, and at least 1,500 species are currently recognized. They are estimated to constitut ...s; some bacteria are also osmophilic. Osmophilic yeasts are important because they cause spoilage in the sugar and sweet goods industry, with products such as fruit juices, fruit juice concentrates, liquid sug ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Probiotics
Probiotics are live microorganisms promoted with claims that they provide health benefits when consumed, generally by improving or restoring the gut microbiota. Probiotics are considered generally safe to consume, but may cause bacteria-host interactions and unwanted side effects in rare cases. There is some evidence that probiotics are beneficial for some conditions, but there is little evidence for many of the health benefits claimed for them. The first discovered probiotic was a certain strain of bacillus in Bulgarian yoghurt, called ''Lactobacillus bulgaricus''. The discovery was made in 1905 by Bulgarian physician and microbiologist Stamen Grigorov. The modern-day theory is generally attributed to Russian Nobel laureate Élie Metchnikoff, who postulated around 1907 that yoghurt-consuming Bulgarian peasants lived longer. A growing probiotics market has led to the need for stricter requirements for scientific substantiation of putative benefits conferred by microorganisms ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |