|
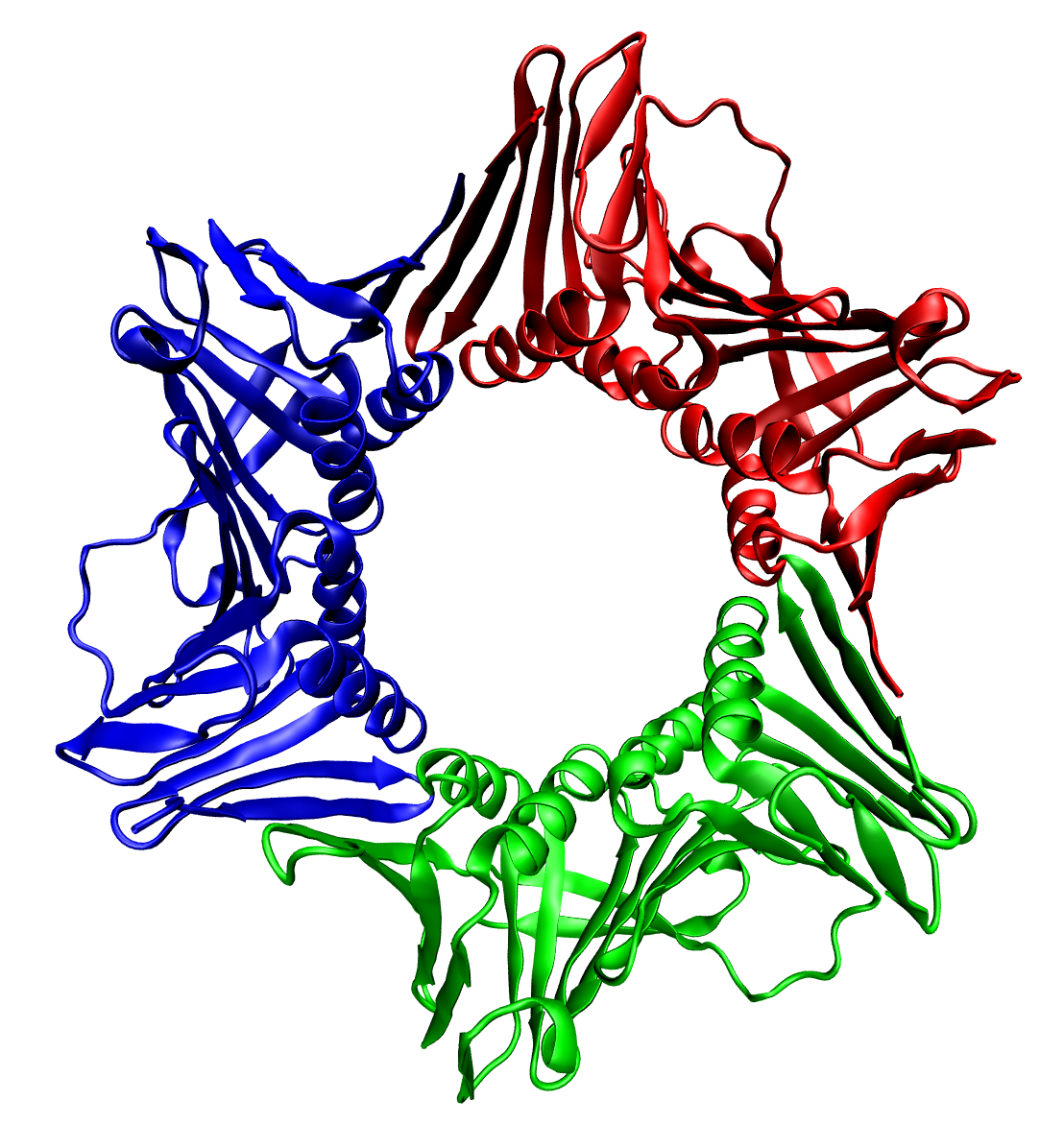
GPCR Oligomer
A GPCR oligomer is a protein complex that consists of a small number ( ''oligoi'' "a few", ''méros'' "part, piece, component") of G protein-coupled receptors (GPCRs). It is held together by covalent bonds or by intermolecular forces. The subunits within this complex are called protomers, while unconnected receptors are called monomers. Receptor homomers consist of identical protomers, while heteromers consist of different protomers. Receptor homodimers – which consist of two identical GPCRs – are the simplest homomeric GPCR oligomers. Receptor heterodimers – which consist of two different GPCRs – are the simplest heteromeric GPCR oligomers. The existence of receptor oligomers is a general phenomenon, whose discovery has superseded the prevailing paradigmatic concept of the function of receptors as plain monomers, and has far-reaching implications for the understanding of neurobiological diseases as well as for the development of drugs. Discovery For a long time it ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
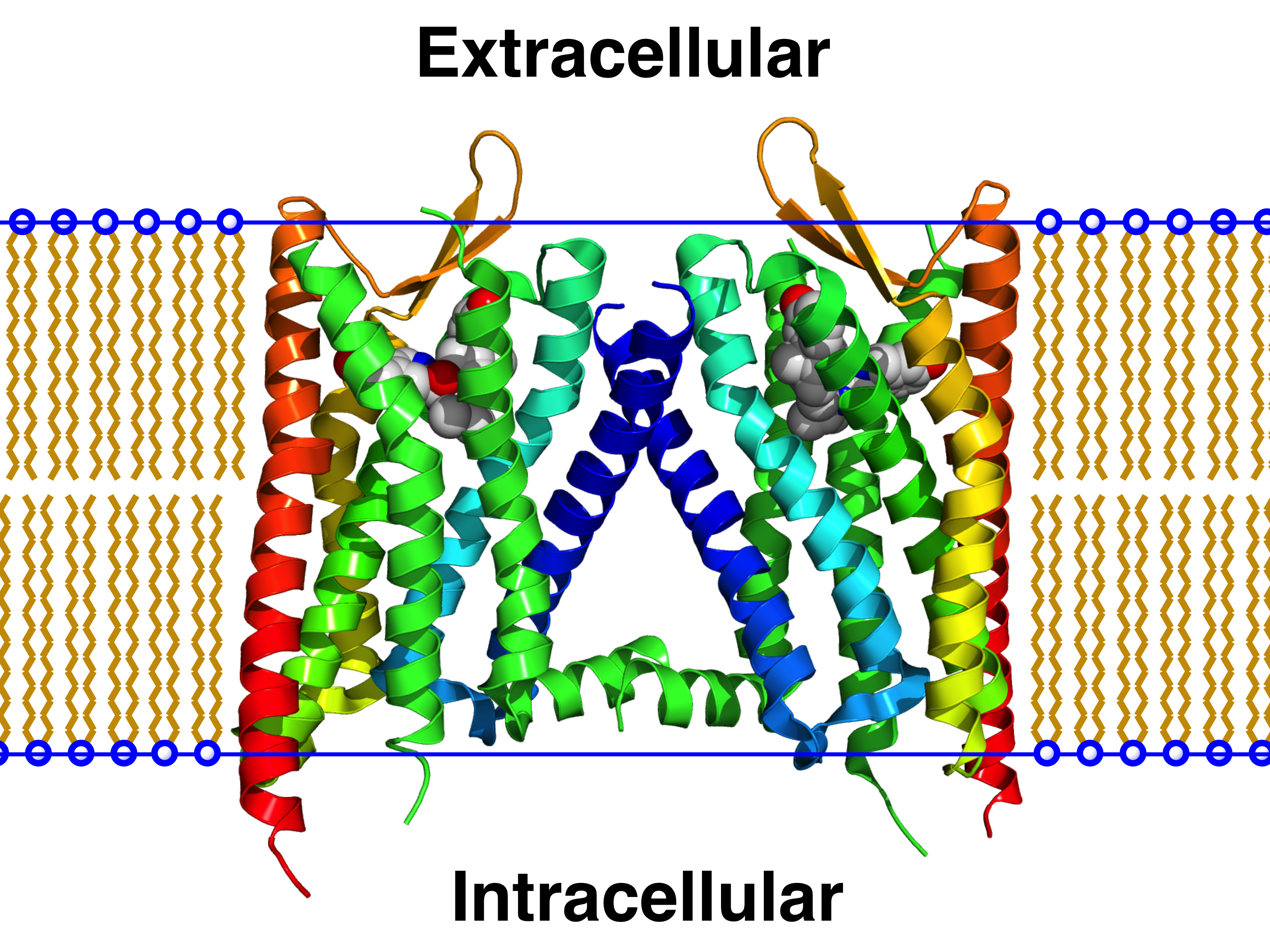
Adenosine A2A Receptor
The adenosine A2A receptor, also known as ADORA2A, is an adenosine receptor, and also denotes the human gene encoding it. Structure This protein is a member of the G protein-coupled receptor (GPCR) family which possess seven transmembrane alpha helices, as well as an extracellular N-terminus and an intracellular C-terminus. Furthermore, located in the intracellular side close to the membrane is a small alpha helix, often referred to as helix 8 (H8). The crystallographic structure of the adenosine A2A receptor reveals a ligand binding pocket distinct from that of other structurally determined GPCRs (i.e., the beta-2 adrenergic receptor and rhodopsin).; Below this primary ( orthosteric) binding pocket lies a secondary ( allosteric) binding pocket. The crystal-structure of A2A bound to the antagonist ZM241385 (PDB code: 4EIY) showed that a sodium-ion can be found in this location of the protein, thus giving it the name 'sodium-ion binding pocket'. Heteromers The actions of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Binding Site
In biochemistry and molecular biology, a binding site is a region on a macromolecule such as a protein that binds to another molecule with specificity. The binding partner of the macromolecule is often referred to as a ligand. Ligands may include other proteins (resulting in a protein-protein interaction), enzyme substrates, second messengers, hormones, or allosteric modulators. The binding event is often, but not always, accompanied by a conformational change that alters the protein's function. Binding to protein binding sites is most often reversible (transient and non-covalent), but can also be covalent reversible or irreversible. Function Binding of a ligand to a binding site on protein often triggers a change in conformation in the protein and results in altered cellular function. Hence binding site on protein are critical parts of signal transduction pathways. Types of ligands include neurotransmitters, toxins, neuropeptides, and steroid hormones. Binding sites in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biomolecular Structure
Biomolecular structure is the intricate folded, three-dimensional shape that is formed by a molecule of protein, DNA, or RNA, and that is important to its function. The structure of these molecules may be considered at any of several length scales ranging from the level of individual atoms to the relationships among entire protein subunits. This useful distinction among scales is often expressed as a decomposition of molecular structure into four levels: primary, secondary, tertiary, and quaternary. The scaffold for this multiscale organization of the molecule arises at the secondary level, where the fundamental structural elements are the molecule's various hydrogen bonds. This leads to several recognizable ''domains'' of protein structure and nucleic acid structure, including such secondary-structure features as alpha helixes and beta sheets for proteins, and hairpin loops, bulges, and internal loops for nucleic acids. The terms ''primary'', ''secondary'', ''tertiary'', and '' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Tetramers
A tetrameric protein is a protein with a quaternary structure of four subunits (tetrameric). Homotetramers have four identical subunits (such as glutathione S-transferase), and heterotetramers are complexes of different subunits. A tetramer can be assembled as dimer of dimers with two homodimer subunits (such as sorbitol dehydrogenase), or two heterodimer subunits (such as hemoglobin). Subunit interactions in tetramers The interactions between subunits forming a tetramer is primarily determined by non covalent interaction. Hydrophobic effects, hydrogen bonds and electrostatic interactions are the primary sources for this binding process between subunits. For homotetrameric proteins such as Sorbitol dehydrogenase (SDH), the structure is believed to have evolved going from a monomeric to a dimeric and finally a tetrameric structure in evolution. The binding process in SDH and many other tetrameric enzymes can be described by the gain in free energy which can be determined from ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Trimer
In biochemistry, a protein trimer is a macromolecular complex formed by three, usually non-covalently bound, macromolecules like proteins or nucleic acids. A homotrimer would be formed by three identical molecules. A heterotrimer would be formed by three different macromolecules. Type II Collagen is an example of homotrimeric protein. Porins usually arrange themselves in membranes as trimers. Bacteriophage T4 tail fiber Multiple copies of a polypeptide encoded by a gene often can form an aggregate referred to as a multimer. When a multimer is formed from polypeptides produced by two different mutant alleles of a particular gene, the mixed multimer may exhibit greater functional activity than the unmixed multimers formed by each of the mutants alone. When a mixed multimer displays increased functionality relative to the unmixed multimers, the phenomenon is referred to as intragenic complementation. The distal portion of each of the bacteriophage T4 tail fibers is encoded by ge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Dimer
In biochemistry, a protein dimer is a macromolecular complex formed by two protein monomers, or single proteins, which are usually non-covalently bound. Many macromolecules, such as proteins or nucleic acids, form dimers. The word ''dimer'' has roots meaning "two parts", '' di-'' + '' -mer''. A protein dimer is a type of protein quaternary structure. A protein homodimer is formed by two identical proteins. A protein heterodimer is formed by two different proteins. Most protein dimers in biochemistry are not connected by covalent bonds. An example of a non-covalent heterodimer is the enzyme reverse transcriptase, which is composed of two different amino acid chains. An exception is dimers that are linked by disulfide bridges such as the homodimeric protein NEMO. Some proteins contain specialized domains to ensure dimerization (dimerization domains) and specificity. The G protein-coupled cannabinoid receptors have the ability to form both homo- and heterodimers with several ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CXCR4
C-X-C chemokine receptor type 4 (CXCR-4) also known as fusin or CD184 (cluster of differentiation 184) is a protein that in humans is encoded by the ''CXCR4'' gene. The protein is a CXC chemokine receptor. Function CXCR-4 is an alpha-chemokine receptor specific for stromal-derived-factor-1 ( SDF-1 also called CXCL12), a molecule endowed with potent chemotactic activity for lymphocytes. CXCR4 is one of several chemokine co-receptors that HIV can use to infect CD4+ T cells. HIV isolates that use CXCR4 are traditionally known as T-cell tropic isolates. Typically, these viruses are found late in infection. It is unclear as to whether the emergence of CXCR4-using HIV is a consequence or a cause of immunodeficiency. CXCR4 is upregulated during the implantation window in natural and hormone replacement therapy cycles in the endometrium, producing, in presence of a human blastocyst, a surface polarization of the CXCR4 receptors suggesting that this receptor is implicated in t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Saccharomyces Cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungus microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have been originally isolated from the skin of grapes. It is one of the most intensively studied eukaryotic model organisms in molecular biology, molecular and cell biology, much like ''Escherichia coli'' as the model bacteria, bacterium. It is the microorganism behind the most common type of fermentation (biochemistry), fermentation. ''S. cerevisiae'' cells are round to ovoid, 5–10 micrometre, μm in diameter. It reproduces by budding. Many proteins important in human biology were first discovered by studying their Homology (biology), homologs in yeast; these proteins include cell cycle proteins, signaling proteins, and protein-processing enzymes. ''S. cerevisiae'' is currently the only yeast cell known to have Berkeley body, Berkeley bo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Förster Resonance Energy Transfer
Förster resonance energy transfer (FRET), fluorescence resonance energy transfer, resonance energy transfer (RET) or electronic energy transfer (EET) is a mechanism describing energy transfer between two light-sensitive molecules ( chromophores). A donor chromophore, initially in its electronic excited state, may transfer energy to an acceptor chromophore through nonradiative dipole–dipole coupling. The efficiency of this energy transfer is inversely proportional to the sixth power of the distance between donor and acceptor, making FRET extremely sensitive to small changes in distance. Measurements of FRET efficiency can be used to determine if two fluorophores are within a certain distance of each other. Such measurements are used as a research tool in fields including biology and chemistry. FRET is analogous to near-field communication, in that the radius of interaction is much smaller than the wavelength of light emitted. In the near-field region, the excited chromophore e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha-2C Adrenergic Receptor
The alpha-2C adrenergic receptor (α2C adrenoceptor), also known as ADRA2C, is an alpha-2 adrenergic receptor, and also denotes the human gene encoding it. Receptor Alpha-2-adrenergic receptors include 3 highly homologous subtypes: alpha2A, alpha2B, and alpha2C. These receptors have a critical role in regulating neurotransmitter release from sympathetic nerves and from adrenergic neurons in the central nervous system. Studies in mice revealed that both the alpha2A and alpha2C subtypes were required for normal presynaptic control of transmitter release from sympathetic nerves in the heart and from central noradrenergic neurons; the alpha2A subtype inhibited transmitter release at high stimulation frequencies, whereas the alpha2C subtype modulated neurotransmission at lower levels of nerve activity. Gene This gene encodes the alpha2C subtype, which contains no introns in either its coding or untranslated sequences. Ligands Agonists * (R)-3-Nitrobiphenyline (also weak ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Muscarinic Acetylcholine Receptor M3
The muscarinic acetylcholine receptor, also known as cholinergic/acetylcholine receptor M3, or the muscarinic 3, is a muscarinic acetylcholine receptor encoded by the human gene CHRM3. The M3 muscarinic receptors are located at many places in the body, e.g., smooth muscles, the endocrine glands, the exocrine glands, lungs, pancreas and the brain. In the CNS, they induce emesis. Muscarinic M3 receptors are expressed in regions of the brain that regulate insulin homeostasis, such as the hypothalamus and dorsal vagal complex of the brainstem. These receptors are highly expressed on pancreatic beta cells and are critical regulators of glucose homoestasis by modulating insulin secretion. In general, they cause smooth muscle contraction and increased glandular secretions. They are unresponsive to PTX and CTX. Mechanism Like the M1 muscarinic receptor, M3 receptors are coupled to G proteins of class Gq, which upregulate phospholipase C and, therefore, inositol trisphosphate and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |