|
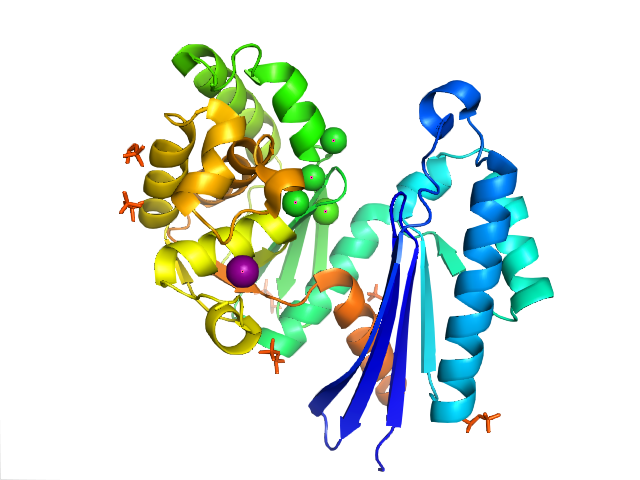
Exopolyphosphatase
Exopolyphosphatase (PPX) is a phosphatase enzyme which catalyzes the hydrolysis of inorganic polyphosphate, a linear molecule composed of up to 1000 or more monomers linked by phospho-anhydride bonds. PPX is a processive exophosphatase, which means that it begins at the ends of the polyphosphate chain and cleaves the phospho-anhydride bonds to release orthophosphate as it moves along the polyphosphate molecule. PPX has several characteristics which distinguish it from other known polyphosphatases, namely that it does not act on ATP, has a strong preference for long chain polyphosphate, and has a very low affinity for polyphosphate molecules with less than 15 phosphate monomers. PPX plays an important role in the metabolism of phosphate and energy in all living organisms. It is especially important for maintenance of appropriate levels of intracellular polyphosphate, which has been implicated in a variety of cellular functions including response to stressors such as deficiencie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphatase
In biochemistry, a phosphatase is an enzyme that uses water to cleave a phosphoric acid Ester, monoester into a phosphate ion and an Alcohol (chemistry), alcohol. Because a phosphatase enzyme catalysis, catalyzes the hydrolysis of its Substrate (chemistry), substrate, it is a subcategory of hydrolases. Phosphatase enzymes are essential to many biological functions, because phosphorylation (e.g. by protein kinases) and dephosphorylation (by phosphatases) serve diverse roles in cell growth, cellular regulation and cell signaling, signaling. Whereas phosphatases remove phosphate groups from molecules, kinases catalyze the transfer of phosphate groups to molecules from Adenosine triphosphate, ATP. Together, kinases and phosphatases direct a form of post-translational modification that is essential to the cell's regulatory network. Phosphatase enzymes are not to be confused with phosphorylase enzymes, which catalyze the transfer of a phosphate group from hydrogen phosphate to an acce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
In Vitro
''In vitro'' (meaning in glass, or ''in the glass'') studies are performed with microorganisms, cells, or biological molecules outside their normal biological context. Colloquially called "test-tube experiments", these studies in biology and its subdisciplines are traditionally done in labware such as test tubes, flasks, Petri dishes, and microtiter plates. Studies conducted using components of an organism that have been isolated from their usual biological surroundings permit a more detailed or more convenient analysis than can be done with whole organisms; however, results obtained from ''in vitro'' experiments may not fully or accurately predict the effects on a whole organism. In contrast to ''in vitro'' experiments, ''in vivo'' studies are those conducted in living organisms, including humans, and whole plants. Definition ''In vitro'' ( la, in glass; often not italicized in English usage) studies are conducted using components of an organism that have been isolated fro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules, such as proteins and nucleic acids. The data, typically obtained by X-ray crystallography, NMR spectroscopy, or, increasingly, cryo-electron microscopy, and submitted by biologists and biochemists from around the world, are freely accessible on the Internet via the websites of its member organisations (PDBe, PDBj, RCSB, and BMRB). The PDB is overseen by an organization called the Worldwide Protein Data Bank, wwPDB. The PDB is a key in areas of structural biology, such as structural genomics. Most major scientific journals and some funding agencies now require scientists to submit their structure data to the PDB. Many other databases use protein structures deposited in the PDB. For example, SCOP and CATH classify protein structures, while PDBsum provides a graphic overview of PDB entries using information from other sources, such as Gene ontology. History Two force ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monomer
In chemistry, a monomer ( ; ''mono-'', "one" + '' -mer'', "part") is a molecule that can react together with other monomer molecules to form a larger polymer chain or three-dimensional network in a process called polymerization. Classification Monomers can be classified in many ways. They can be subdivided into two broad classes, depending on the kind of the polymer that they form. Monomers that participate in condensation polymerization have a different stoichiometry than monomers that participate in addition polymerization: : Other classifications include: *natural vs synthetic monomers, e.g. glycine vs caprolactam, respectively *polar vs nonpolar monomers, e.g. vinyl acetate vs ethylene, respectively *cyclic vs linear, e.g. ethylene oxide vs ethylene glycol, respectively The polymerization of one kind of monomer gives a homopolymer. Many polymers are copolymers, meaning that they are derived from two different monomers. In the case of condensation polymerizations, the r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dimer (chemistry)
A dimer () ('' di-'', "two" + ''-mer'', "parts") is an oligomer consisting of two monomers joined by bonds that can be either strong or weak, covalent or intermolecular. Dimers also have significant implications in polymer chemistry, inorganic chemistry, and biochemistry. The term ''homodimer'' is used when the two molecules are identical (e.g. A–A) and ''heterodimer'' when they are not (e.g. A–B). The reverse of dimerization is often called dissociation. When two oppositely charged ions associate into dimers, they are referred to as ''Bjerrum pairs'', after Niels Bjerrum. Noncovalent dimers Anhydrous carboxylic acids form dimers by hydrogen bonding of the acidic hydrogen and the carbonyl oxygen. For example, acetic acid forms a dimer in the gas phase, where the monomer units are held together by hydrogen bonds. Under special conditions, most OH-containing molecules form dimers, e.g. the water dimer. Excimers and exciplexes are excited structures with a short lifetime. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Active Site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate (binding site) and residues that catalyse a reaction of that substrate (catalytic site). Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes. Each active site is evolved to be optimised to bind a particular substrate and catalyse a particular reaction, resulting in high specificity. This specificity is determined by the arrangement of amino acids within the active site and the structure of the substrates. Sometimes enzymes also need to bind with some cofactors to fulfil their function. The active si ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha-helices
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astbur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antiparallel (biochemistry)
In biochemistry, two biopolymers are antiparallel if they run parallel to each other but with opposite directionality (alignments). An example is the two complementary strands of a DNA double helix, which run in opposite directions alongside each other. Nucleic acids Nucleic acid molecules have a phosphoryl (5') end and a hydroxyl (3') end. This notation follows from organic chemistry nomenclature, and can be used to define the movement of enzymes such as DNA polymerases relative to the DNA strand in a non-arbitrary manner. G-quadruplexes G-quadruplexes, also known as G4 DNA are secondary structures found in nucleic acids that are rich in guanine. These structures are normally located at the telomeres (the ends of the chromosomes). The G-quadruplex can either be parallel or antiparallel depending on the loop configuration, which is a component of the structure. If all the DNA strands run in the same direction, it is termed to be a parallel quadruplex, and is known as a str ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Sheet
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, notably Alzheimer's disease. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. A refined versi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribonuclease
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 (for the phosphorolytic enzymes) and 3.1 (for the hydrolytic enzymes) classes of enzymes. Function All organisms studied contain many RNases of two different classes, showing that RNA degradation is a very ancient and important process. As well as clearing of cellular RNA that is no longer required, RNases play key roles in the maturation of all RNA molecules, both messenger RNAs that carry genetic material for making proteins and non-coding RNAs that function in varied cellular processes. In addition, active RNA degradation systems are the first defense against RNA viruses and provide the underlying machinery for more advanced cellular immune strategies such as RNAi. Some cells also secrete copious quantities of non-specific RN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aquifex Aeolicus
"''Aquifex aeolicus''" is a chemolithoautotrophic, Gram-negative, motile, hyperthermophilic bacterium. "''A. aeolicus"'' is generally rod-shaped with an approximate length of 2.0-6.0μm and a diameter of 0.4-0.5μm. "''A. aeolicus''" is neither validly nor effectively published and, having no standing in nomenclature, should be styled in quotation marks. It is one of a handful of species in the Aquificota phylum, an unusual group of thermophilic bacteria that are thought to be some of the oldest species of bacteria, related to filamentous bacteria first observed at the turn of the century. "''A. aeolicus''" is also believed to be one of the earliest diverging species of thermophilic bacteria. "''A. aeolicus''" grows best in water between 85 °C and 95 °C, and can be found near underwater volcanoes or hot springs. It requires oxygen to survive but has been found to grow optimally under microaerophilic conditions. Due to its high stability against high temperature and la ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |





_structure.png)
