|
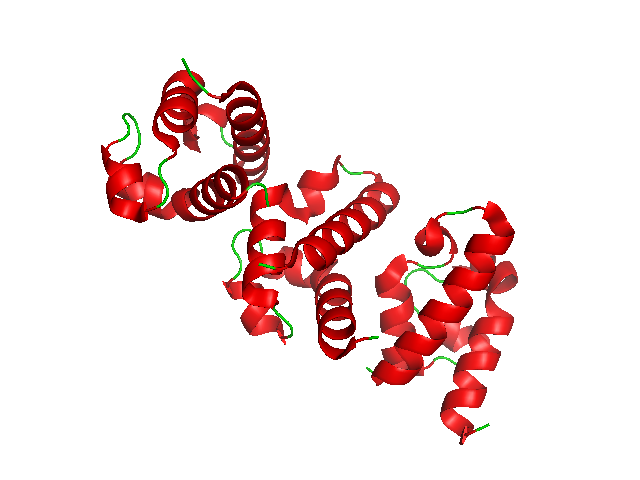
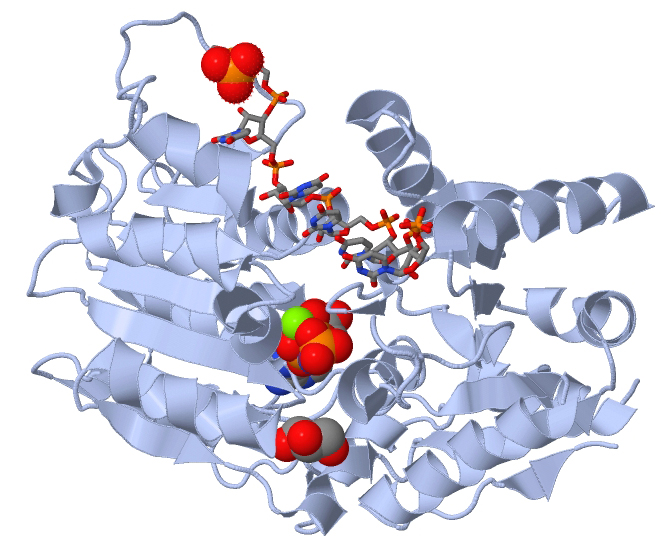
EIF4A Helicase
Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two hybridized nucleic acid strands (hence '' helic- + -ase''), using energy from ATP hydrolysis. There are many helicases, representing the great variety of processes in which strand separation must be catalyzed. Approximately 1% of eukaryotic genes code for helicases. The human genome codes for 95 non-redundant helicases: 64 RNA helicases and 31 DNA helicases. Many cellular processes, such as DNA replication, transcription, translation, recombination, DNA repair, and ribosome biogenesis involve the separation of nucleic acid strands that necessitates the use of helicases. Some specialized helicases are also involved in sensing of viral nucleic acids during infection and fulfill a immunological function. Function Helicases ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Helicase
Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two hybridized nucleic acid strands (hence '' helic- + -ase''), using energy from ATP hydrolysis. There are many helicases, representing the great variety of processes in which strand separation must be catalyzed. Approximately 1% of eukaryotic genes code for helicases. The human genome codes for 95 non-redundant helicases: 64 RNA helicases and 31 DNA helicases. Many cellular processes, such as DNA replication, transcription, translation, recombination, DNA repair, and ribosome biogenesis involve the separation of nucleic acid strands that necessitates the use of helicases. Some specialized helicases are also involved in sensing of viral nucleic acids during infection and fulfill a immunological function. Function Helicases are o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
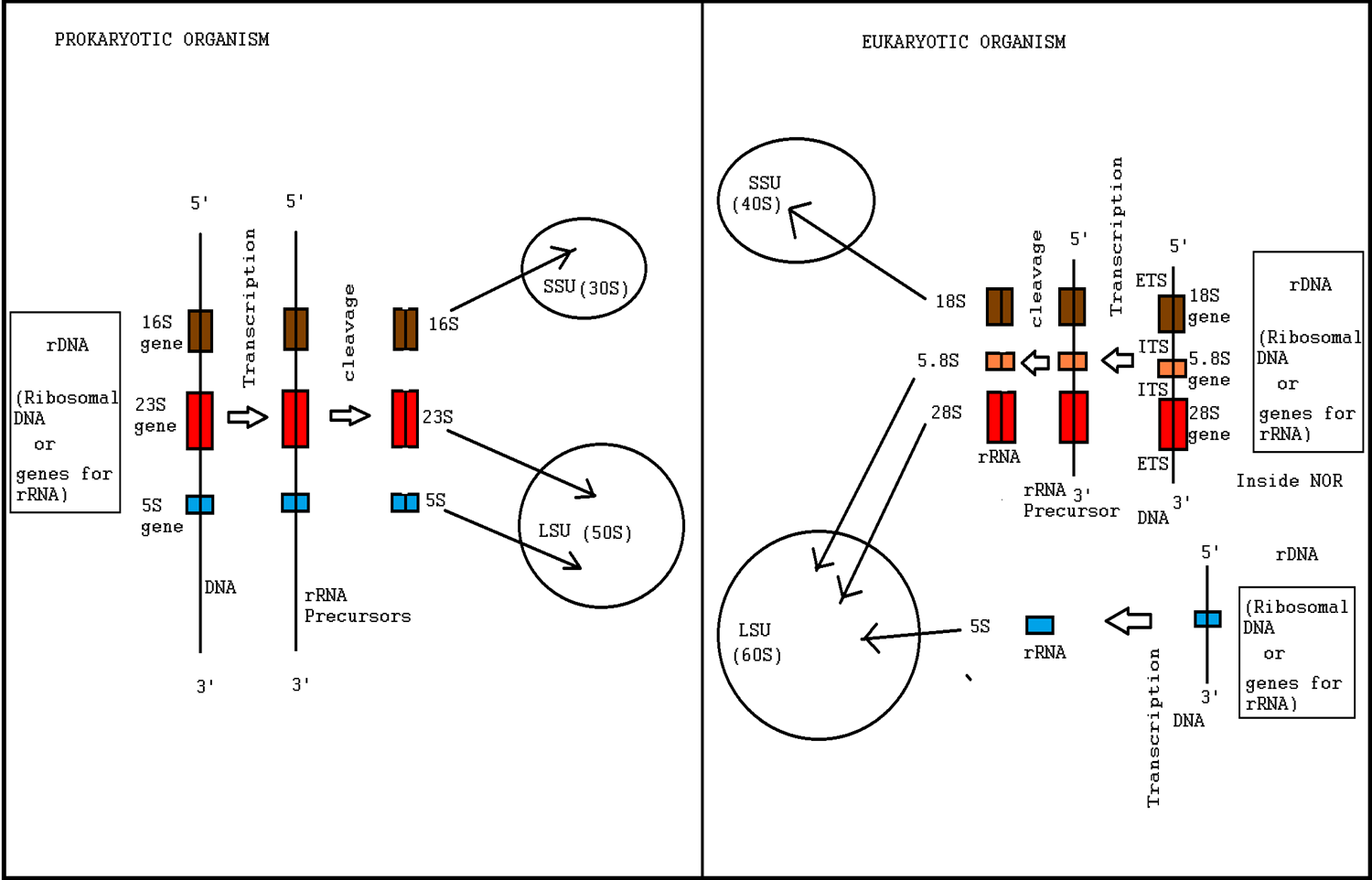
Ribosome Biogenesis
Ribosome biogenesis is the process of making ribosomes. In prokaryotes, this process takes place in the cytoplasm with the transcription of many ribosome gene operons. In eukaryotes, it takes place both in the cytoplasm and in the nucleolus. It involves the coordinated function of over 200 proteins in the synthesis and processing of the three prokaryotic or four eukaryotic rRNAs, as well as assembly of those rRNAs with the ribosomal proteins. Most of the ribosomal proteins fall into various energy-consuming enzyme families including ATP-dependent RNA helicases, AAA-ATPases, GTPases, and kinases. About 60% of a cell's energy is spent on ribosome production and maintenance. Ribosome biogenesis is a very tightly regulated process, and it is closely linked to other cellular activities like growth and division. Some have speculated that in the origin of life, ribosome biogenesis predates cells, and that genes and cells evolved to enhance the reproductive capacity of ribosomes. R ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
In Vivo
Studies that are ''in vivo'' (Latin for "within the living"; often not italicized in English) are those in which the effects of various biological entities are tested on whole, living organisms or cells, usually animals, including humans, and plants, as opposed to a tissue extract or dead organism. This is not to be confused with experiments done ''in vitro'' ("within the glass"), i.e., in a laboratory environment using test tubes, Petri dishes, etc. Examples of investigations ''in vivo'' include: the pathogenesis of disease by comparing the effects of bacterial infection with the effects of purified bacterial toxins; the development of non-antibiotics, antiviral drugs, and new drugs generally; and new surgical procedures. Consequently, animal testing and clinical trials are major elements of ''in vivo'' research. ''In vivo'' testing is often employed over ''in vitro'' because it is better suited for observing the overall effects of an experiment on a living subject. In dr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Dimer
In biochemistry, a protein dimer is a macromolecular complex formed by two protein monomers, or single proteins, which are usually non-covalently bound. Many macromolecules, such as proteins or nucleic acids, form dimers. The word ''dimer'' has roots meaning "two parts", '' di-'' + '' -mer''. A protein dimer is a type of protein quaternary structure. A protein homodimer is formed by two identical proteins. A protein heterodimer is formed by two different proteins. Most protein dimers in biochemistry are not connected by covalent bonds. An example of a non-covalent heterodimer is the enzyme reverse transcriptase, which is composed of two different amino acid chains. An exception is dimers that are linked by disulfide bridges such as the homodimeric protein NEMO. Some proteins contain specialized domains to ensure dimerization (dimerization domains) and specificity. The G protein-coupled cannabinoid receptors have the ability to form both homo- and heterodimers with several ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monomer
In chemistry, a monomer ( ; ''mono-'', "one" + '' -mer'', "part") is a molecule that can react together with other monomer molecules to form a larger polymer chain or three-dimensional network in a process called polymerization. Classification Monomers can be classified in many ways. They can be subdivided into two broad classes, depending on the kind of the polymer that they form. Monomers that participate in condensation polymerization have a different stoichiometry than monomers that participate in addition polymerization: : Other classifications include: *natural vs synthetic monomers, e.g. glycine vs caprolactam, respectively *polar vs nonpolar monomers, e.g. vinyl acetate vs ethylene, respectively *cyclic vs linear, e.g. ethylene oxide vs ethylene glycol, respectively The polymerization of one kind of monomer gives a homopolymer. Many polymers are copolymers, meaning that they are derived from two different monomers. In the case of condensation polymerizations, the r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hexamer
In chemistry and biochemistry, an oligomer () is a molecule that consists of a few repeating units which could be derived, actually or conceptually, from smaller molecules, monomers.Quote: ''Oligomer molecule: A molecule of intermediate relative molecular mass, the structure of which essentially comprises a small plurality of units derived, actually or conceptually, from molecules of lower relative molecular mass.'' The name is composed of Greek elements '' oligo-'', "a few" and '' -mer'', "parts". An adjective form is ''oligomeric''. The oligomer concept is contrasted to that of a polymer, which is usually understood to have a large number of units, possibly thousands or millions. However, there is no sharp distinction between these two concepts. One proposed criterion is whether the molecule's properties vary significantly with the removal of one or a few of the units. An oligomer with a specific number of units is referred to by the Greek prefix denoting that number, wit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DnaB Helicase
DnaB helicase is an enzyme in bacteria which opens the replication fork during DNA replication. Although the mechanism by which DnaB both couples ATP hydrolysis to translocation along DNA and denatures the duplex is unknown, a change in the quaternary structure of the protein involving dimerisation of the N-terminal domain has been observed and may occur during the enzymatic cycle. Initially when DnaB binds to dnaA, it is associated with dnaC, a negative regulator. After DnaC dissociates, DnaB binds dnaG. The N-terminal has a multi-helical structure that forms an orthogonal bundle. The C-terminal domain contains an ATP-binding site and is therefore probably the site of ATP hydrolysis. In eukaryotes, helicase function is provided by the MCM (Minichromosome maintenance) complex. The DnaB helicase is the product of the ''dnaB'' gene. The helicase enzyme that is produced is a hexamer in ''E. coli'', as well as in many other bacteria. The energy for DnaB activity is provided by N ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligomerization
In chemistry and biochemistry, an oligomer () is a molecule that consists of a few repeating units which could be derived, actually or conceptually, from smaller molecules, monomers.Quote: ''Oligomer molecule: A molecule of intermediate relative molecular mass, the structure of which essentially comprises a small plurality of units derived, actually or conceptually, from molecules of lower relative molecular mass.'' The name is composed of Greek elements '' oligo-'', "a few" and '' -mer'', "parts". An adjective form is ''oligomeric''. The oligomer concept is contrasted to that of a polymer, which is usually understood to have a large number of units, possibly thousands or millions. However, there is no sharp distinction between these two concepts. One proposed criterion is whether the molecule's properties vary significantly with the removal of one or a few of the units. An oligomer with a specific number of units is referred to by the Greek prefix denoting that number, wit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Processivity
In molecular biology and biochemistry, processivity is an enzyme's ability to catalyze "consecutive reactions without releasing its substrate". For example, processivity is the average number of nucleotides added by a polymerase enzyme, such as DNA polymerase, per association event with the template strand. Because the binding of the polymerase to the template is the rate-limiting step in DNA synthesis, the overall rate of DNA replication during S phase of the cell cycle is dependent on the processivity of the DNA polymerases performing the replication. DNA clamp proteins are integral components of the DNA replication machinery and serve to increase the processivity of their associated polymerases. Some polymerases add over 50,000 nucleotides to a growing DNA strand before dissociating from the template strand, giving a replication rate of up to 1,000 nucleotides per second. DNA binding interactions Polymerases interact with the phosphate backbone and the minor groove of the DNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Directionality (molecular Biology)
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide pentose-sugar-ring means that there will be a 5′ end (usually pronounced "five-prime end"), which frequently contains a phosphate group attached to the 5′ carbon of the ribose ring, and a 3′ end (usually pronounced "three-prime end"), which typically is unmodified from the ribose -OH substituent. In a DNA double helix, the strands run in opposite directions to permit base pairing between them, which is essential for replication or transcription of the encoded information. Nucleic acids can only be synthesized in vivo in the 5′-to-3′ direction, as the polymerases that assemble various types of new strands generally rely on the energy produced by breaking nucleoside triphosphate bonds to attach new nucleoside monophosphates to the 3′- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Editing
RNA editing (also RNA modification) is a molecular process through which some cells can make discrete changes to specific nucleotide sequences within an RNA molecule after it has been generated by RNA polymerase. It occurs in all living organisms and is one of the most evolutionarily conserved properties of RNAs. RNA editing may include the insertion, deletion, and base substitution of nucleotides within the RNA molecule. RNA editing is relatively rare, with common forms of RNA processing (e.g. splicing, 5'-capping, and 3'-polyadenylation) not usually considered as editing. It can affect the activity, localization as well as stability of RNAs, and has been linked with human diseases. RNA editing has been observed in some tRNA, rRNA, mRNA, or miRNA molecules of eukaryotes and their viruses, archaea, and prokaryotes. RNA editing occurs in the cell nucleus, as well as within mitochondria and plastids. In vertebrates, editing is rare and usually consists of a small number of changes t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Splicing
RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcript is transformed into a mature messenger RNA (mRNA). It works by removing all the introns (non-coding regions of RNA) and ''splicing'' back together exons (coding regions). For nuclear-encoded genes, splicing occurs in the nucleus either during or immediately after transcription. For those eukaryotic genes that contain introns, splicing is usually needed to create an mRNA molecule that can be translated into protein. For many eukaryotic introns, splicing occurs in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs). There exist self-splicing introns, that is, ribozymes that can catalyze their own excision from their parent RNA molecule. The process of transcription, splicing and translation is called gene expression, the central dogma of molecular biology. Splicing pathways Several methods of RNA splici ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |