|
EF2
Elongation factors are a set of proteins that function at the ribosome, during protein synthesis, to facilitate translational elongation from the formation of the first to the last peptide bond of a growing polypeptide. Most common elongation factors in prokaryotes are EF-Tu, EF-Ts, EF-G. Bacteria and eukaryotes use elongation factors that are largely homologous to each other, but with distinct structures and different research nomenclatures. Elongation is the most rapid step in translation. In bacteria, it proceeds at a rate of 15 to 20 amino acids added per second (about 45-60 nucleotides per second). In eukaryotes the rate is about two amino acids per second (about 6 nucleotides read per second). Elongation factors play a role in orchestrating the events of this process, and in ensuring the high accuracy translation at these speeds. Nomenclature of homologous EFs In addition to their cytoplasmic machinery, eukaryotic mitochondria and plastids have their own translation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
EF-G
EF-G (elongation factor G, historically known as translocase) is a prokaryotic elongation factor involved in mRNA translation. As a GTPase, EF-G catalyzes the movement (translocation) of transfer RNA (tRNA) and messenger RNA (mRNA) through the ribosome. Structure Encoded by the ''fusA'' gene on the ''str'' operon, EF-G is made up of 704 amino acids that form 5 domains, labeled Domain I through Domain V. Domain I may be referred to as the G-domain or as Domain I(G), since it binds to and hydrolyzes guanosine triphosphate (GTP). Domain I also helps EF-G bind to the ribosome, and contains the N-terminal of the polypeptide chain. Domain IV is important for translocation, as it undergoes a significant conformational change and enters the A site on the 30S ribosomal subunit, pushing the mRNA and tRNA molecules from the A site to the P site. The five domains may be also separated into two super-domains. Super-domain I consists of Domains I and II, and super-domain II consists of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
EEF-2
Eukaryotic elongation factor 2 is a protein that in humans is encoded by the ''EEF2'' gene. It is the archaeal and eukaryotic counterpart of bacterial EF-G. This gene encodes a member of the GTP-binding translation elongation factor family. This protein is an essential factor for protein synthesis. It promotes the GTP-dependent translocation of the ribosome. This protein is completely inactivated by EF-2 kinase phosphorylation. aEF2/eEF2 found in most archaea and eukaryotes, including humans, contains a post translationally modified histidine diphthamide. It is the target of diphtheria toxin (from ''Corynebacterium diphtheriae''), and exotoxin A (from ''Pseudomonas aeruginosa ''Pseudomonas aeruginosa'' is a common Bacterial capsule, encapsulated, Gram-negative bacteria, Gram-negative, Aerobic organism, aerobic–facultative anaerobe, facultatively anaerobic, Bacillus (shape), rod-shaped bacteria, bacterium that can c ...''). The inactivation of EF-2 by toxins inhibits protei ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Diphtheria Toxin
Diphtheria toxin is an exotoxin secreted mainly by '' Corynebacterium diphtheriae'' but also by ''Corynebacterium ulcerans'' and '' Corynebacterium pseudotuberculosis'', the pathogenic bacterium that causes diphtheria. The toxin gene is encoded by a prophageA prophage is a virus that has inserted itself into the genome of the host bacterium. called corynephage β. The toxin causes the disease in humans by gaining entry into the cell cytoplasm and inhibiting protein synthesis. Structure Diphtheria toxin is a single polypeptide chain of 535 amino acids consisting of two subunits linked by disulfide bridges, known as an A-B toxin. Binding to the cell surface of the B subunit (the less stable of the two subunits) allows the A subunit (the more stable part of the protein) to penetrate the host cell. The crystal structure of the diphtheria toxin homodimer has been determined to 2.5 Ångstrom resolution. The structure reveals a Y-shaped molecule consisting of three domain ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
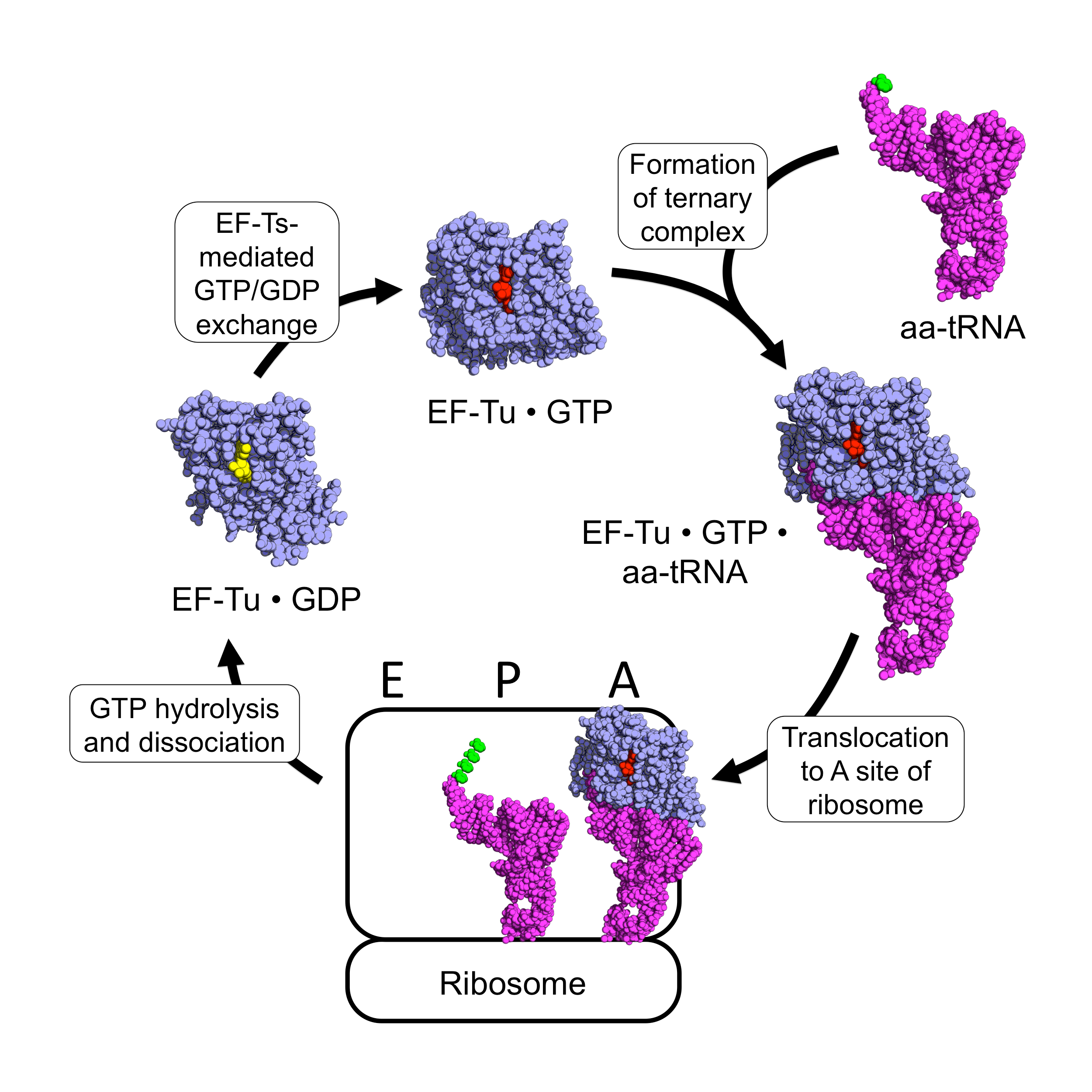
EF-Tu
EF-Tu (elongation factor thermo unstable) is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome. It is a G-protein, and facilitates the selection and binding of an aa-tRNA to the A-site of the ribosome. As a reflection of its crucial role in translation, EF-Tu is one of the most abundant and highly conserved proteins in prokaryotes. It is found in eukaryotic mitochondria as TUFM. As a family of elongation factors, EF-Tu also includes its eukaryotic and archaeal homolog, the alpha subunit of eEF-1 (EF-1A). Background Elongation factors are part of the mechanism that synthesizes new proteins through translation in the ribosome. Transfer RNAs (tRNAs) carry the individual amino acids that become integrated into a protein sequence, and have an anticodon for the specific amino acid that they are charged with. Messenger RNA (mRNA) carries the genetic information that encodes the primary structure of a protein, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
GUF1
Mitochondrial GTP binding elongation factor (''Homo Sapiens'', GUF1), is a protein which in humans is encoded by the ''GUF1'' gene. The GUF1 protein plays an important role in maintaining proper mitochondrial function, ensuring accuracy when mitochondrial genes are being translated. The gene shows the most expression in the brain, while the least expression is found in the pancreas. Function and biochemistry GUF1 is a GTPase that Hydrolysis, hydrolyzes GTP to GDP and plays an important role in maintaining mitochondrial function by ensuring accurate translation of mitochondrial genes. It helps prevent amino acid mis-incorporation under stress by facilitating ribosomal back-translocation during protein synthesis. Additionally, its C-terminal region aids in tRNA interaction, distinguishing it from other GTPases. The highly conserved C-terminal of GUF1 facilitates tRNA binding, while the N-terminal handles GTP binding and hydrolysis. The G domain supports core GTPase activity. Gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Diphtheria
Diphtheria is an infection caused by the bacteria, bacterium ''Corynebacterium diphtheriae''. Most infections are asymptomatic or have a mild Course (medicine), clinical course, but in some outbreaks, the mortality rate approaches 10%. Signs and symptoms may vary from mild to severe, and usually start two to five days after exposure. Symptoms often develop gradually, beginning with a sore throat and fever. In severe cases, a grey or white patch develops in the throat, which can block the airway, and create a barking cough similar to what is observed in croup. The neck may also swell, in part due to the enlargement of the facial lymph nodes. Diphtheria can also involve the skin, eyes, or genitals, and can cause complications, including myocarditis (which in itself can result in an cardiac arrhythmia, abnormal heart rate), peripheral neuropathy, inflammation of nerves (which can result in paralysis), proteinuria, kidney problems, and bleeding problems due to thrombocytopenia, low ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Translation (genetics)
In biology, translation is the process in living cells in which proteins are produced using RNA molecules as templates. The generated protein is a sequence of amino acids. This sequence is determined by the sequence of nucleotides in the RNA. The nucleotides are considered three at a time. Each such triple results in the addition of one specific amino acid to the protein being generated. The matching from nucleotide triple to amino acid is called the genetic code. The translation is performed by a large complex of functional RNA and proteins called ribosomes. The entire process is called gene expression. In translation, messenger RNA (mRNA) is decoded in a ribosome, outside the nucleus, to produce a specific amino acid chain, or polypeptide. The polypeptide later folds into an active protein and performs its functions in the cell. The polypeptide can also start folding during protein synthesis. The ribosome facilitates decoding by inducing the binding of complementary transfe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Selenocysteine
Selenocysteine (symbol Sec or U, in older publications also as Se-Cys) is the 21st proteinogenic amino acid. Selenoproteins contain selenocysteine residues. Selenocysteine is an analogue of the more common cysteine with selenium in place of the sulfur. Selenocysteine is present in several enzymes (for example glutathione peroxidases, tetraiodothyronine 5 deiodinase, tetraiodothyronine 5′ deiodinases, thioredoxin reductases, formate dehydrogenases, glycine reductases, selenophosphate synthetase 2, methionine-''R''-sulfoxide reductase B1 (SEPX1), and some hydrogenases). It occurs in all three Domain (biology), domains of life, including important enzymes (listed above) present in humans. Selenocysteine was discovered in 1974 by biochemist Thressa Stadtman at the National Institutes of Health. Chemistry Selenocysteine is the Se-analogue of cysteine. It is rarely encountered outside of living tissue (nor is it available commercially) because of its high susceptiblility to air-oxi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |