|
EC 5.4.2.10
In enzymology, a phosphoenolpyruvate mutase () is an enzyme that catalyzes the chemical reaction :phosphoenolpyruvate \rightleftharpoons 3-phosphonopyruvate Hence, this enzyme has one substrate, phosphoenolpyruvate (PEP), and one product, 3-phosphonopyruvate (PPR), which are structural isomers. This enzyme belongs to the family of isomerases, specifically the phosphotransferases (phosphomutases), which transfer phosphate groups within a molecule. The systematic name of this enzyme class is phosphoenolpyruvate 2,3-phosphonomutase. Other names in common use include phosphoenolpyruvate-phosphonopyruvate phosphomutase, PEP phosphomutase, phosphoenolpyruvate phosphomutase, PEPPM, and PEP phosphomutase. This enzyme participates in aminophosphonate metabolism. Phosphoenolpyruvate mutase was discovered in 1988. Structural studies As of late 2007, 6 structures have been solved for this class of enzymes, all by the Herzberg grouat the University of Maryland using PEPPM from th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzymology
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Barrel
In protein structures, a beta barrel is a beta sheet composed of tandem repeats that twists and coils to form a closed toroidal structure in which the first strand is bonded to the last strand (hydrogen bond). Beta-strands in many beta-barrels are arranged in an antiparallel fashion. Beta barrel structures are named for resemblance to the barrels used to contain liquids. Most of them are water-soluble proteins and frequently bind hydrophobic ligands in the barrel center, as in lipocalins. Others span cell membranes and are commonly found in porins. Porin-like barrel structures are encoded by as many as 2–3% of the genes in Gram-negative bacteria. It has been shown that more than 600 proteins with various function (e.g., oxidase, dismutase, amylase) contain the beta barrel structure. In many cases, the strands contain alternating polar and non-polar (hydrophilic and hydrophobic) amino acids, so that the hydrophobic residues are oriented into the interior of the barrel to form a hy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Carboxylic Acid
In organic chemistry, a carboxylic acid is an organic acid that contains a carboxyl group () attached to an R-group. The general formula of a carboxylic acid is or , with R referring to the alkyl, alkenyl, aryl, or other group. Carboxylic acids occur widely. Important examples include the amino acids and fatty acids. Deprotonation of a carboxylic acid gives a carboxylate anion. Examples and nomenclature Carboxylic acids are commonly identified by their trivial names. They at oftentimes have the suffix ''-ic acid''. IUPAC-recommended names also exist; in this system, carboxylic acids have an ''-oic acid'' suffix. For example, butyric acid (C3H7CO2H) is butanoic acid by IUPAC guidelines. For nomenclature of complex molecules containing a carboxylic acid, the carboxyl can be considered position one of the parent chain even if there are other substituents, such as 3-chloropropanoic acid. Alternately, it can be named as a "carboxy" or "carboxylic acid" substituent on another ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
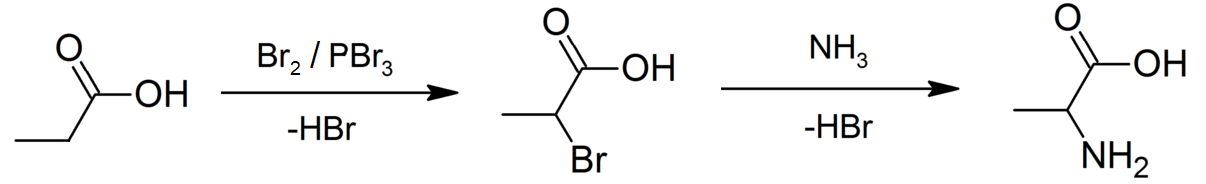
Propanoate
Propionic acid (, from the Greek words πρῶτος : ''prōtos'', meaning "first", and πίων : ''píōn'', meaning "fat"; also known as propanoic acid) is a naturally occurring carboxylic acid with chemical formula CH3CH2CO2H. It is a liquid with a pungent and unpleasant smell somewhat resembling body odor. The anion CH3CH2CO2− as well as the salts and esters of propionic acid are known as propionates or propanoates. History Propionic acid was first described in 1844 by Johann Gottlieb, who found it among the degradation products of sugar. Over the next few years, other chemists produced propionic acid by different means, none of them realizing they were producing the same substance. In 1847, French chemist Jean-Baptiste Dumas established all the acids to be the same compound, which he called propionic acid, from the Greek words πρῶτος (prōtos), meaning ''first'', and πίων (piōn), meaning ''fat'', because it is the smallest H(CH2)''n''COOH acid that exhib ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methylisocitrate Lyase
The enzyme methylisocitrate lyase () catalyzes the chemical reaction :(2''S'',3''R'')-3-hydroxybutane-1,2,3-tricarboxylate \rightleftharpoons pyruvate + succinate The reaction is similar to that of isocitrate lyase, except that an additional methyl group (marked with an asterisk in the above scheme) is present, meaning that citrate is replaced by methylcitrate and glyoxylate by pyruvate. In fact, in some bacteria such as ''Mycobacterium tuberculosis'', isocitrate lyase actually plays the role of methylisocitrate lyase. This enzyme belongs to the family of lyases, specifically the oxo-acid-lyases, which cleave carbon-carbon bonds. The systematic name of this enzyme class is (2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate pyruvate-lyase (succinate-forming). Other names in common use include 2-methylisocitrate lyase, MICL, and (2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate pyruvate-lyase. This enzyme participates in propanoate metabolism. Methylisocitrate lyase was discovered in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Overview PEPPM
Overview may refer to: * Overview article, an artícle that summarizes the current state of understanding on a topic * Overview map, generalised view of a geographic area See also * Summary (other) * Outline (list) * ''A Brief Overview'' * Overview and Scrutiny Overview may refer to: * Overview article, an artícle that summarizes the current state of understanding on a topic * Overview map, generalised view of a geographic area See also * Summary (other) * Outline (list) * ''A Brief Overvie ... * Overview effect * * {{disambiguation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Kinemage
A kinemage (short for kinetic image) is an interactive graphic scientific illustration. It often is used to visualize molecules, especially proteins although it can also represent other types of 3-dimensional data (such as geometric figures, social networks, or tetrahedra oRNA base composition. The kinemage system is designed to optimize ease of use, interactive performance, and the perception and communication of detailed 3D information. The kinemage information is stored in a text file, human- and machine-readable, that describes the hierarchy of display objects and their properties, and includes optional explanatory text. The kinemage format is a defined chemical MIME type of 'chemical/x-kinemage' with the file extension '.kin'. Early history Kinemages were first developed by David Richardson at Duke University School of Medicine, for the Protein Society's journal Protein Science that premiered in January 1992. For its first 5 years (1992–1996), each issue of Protein Scien ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribbon Diagram
Ribbon diagrams, also known as Richardson diagrams, are three-dimensional space, 3D schematic representations of protein structure and are one of the most common methods of protein depiction used today. The ribbon shows the overall path and organization of the protein backbone in 3D, and serves as a visual framework on which to hang details of the full atomic structure, such as the balls for the oxygen atoms bound to the active site of myoglobin in the adjacent image. Ribbon diagrams are generated by interpolating a smooth curve through the polypeptide backbone. Alpha helix, α-helices are shown as coiled ribbons or thick tubes, Beta strand, β-strands as arrows, and non-repetitive coils or loops as lines or thin tubes. The direction of the Peptide, polypeptide chain is shown locally by the arrows, and may be indicated overall by a colour ramp along the length of the ribbon. Ribbon diagrams are simple yet powerful, expressing the visual basics of a molecular structure (twist, fold ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Wild Type
The wild type (WT) is the phenotype of the typical form of a species as it occurs in nature. Originally, the wild type was conceptualized as a product of the standard "normal" allele at a locus, in contrast to that produced by a non-standard, "mutant" allele. "Mutant" alleles can vary to a great extent, and even become the wild type if a genetic shift occurs within the population. Continued advancements in genetic mapping technologies have created a better understanding of how mutations occur and interact with other genes to alter phenotype. It is now appreciated that most or all gene loci exist in a variety of allelic forms, which vary in frequency throughout the geographic range of a species, and that a uniform wild type does not exist. In general, however, the most prevalent allele – i.e., the one with the highest gene frequency – is the one deemed wild type. The concept of wild type is useful in some experimental organisms such as fruit flies ''Drosophila melanogaster'' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conformational Change
In biochemistry, a conformational change is a change in the shape of a macromolecule, often induced by environmental factors. A macromolecule is usually flexible and dynamic. Its shape can change in response to changes in its environment or other factors; each possible shape is called a conformation, and a transition between them is called a ''conformational change''. Factors that may induce such changes include temperature, pH, voltage, light in chromophores, concentration of ions, phosphorylation, or the binding of a ligand. Transitions between these states occur on a variety of length scales (tenths of Å to nm) and time scales (ns to s), and have been linked to functionally relevant phenomena such as allosteric signaling and enzyme catalysis. Laboratory analysis Many biophysical techniques such as crystallography, NMR, electron paramagnetic resonance (EPR) using spin label techniques, circular dichroism (CD), hydrogen exchange, and FRET can be used to study macrom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Active Site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate (binding site) and residues that catalyse a reaction of that substrate (catalytic site). Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes. Each active site is evolved to be optimised to bind a particular substrate and catalyse a particular reaction, resulting in high specificity. This specificity is determined by the arrangement of amino acids within the active site and the structure of the substrates. Sometimes enzymes also need to bind with some cofactors to fulfil their function. The active si ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |