|
Ethylbenzene Dehydrogenase
In enzymology, an ethylbenzene hydroxylase () is an enzyme that catalyzes the chemical reaction :ethylbenzene + H2O + acceptor \rightleftharpoons (S)-1-phenylethanol + reduced acceptor The 3 substrates of this enzyme are ethylbenzene, H2O, and acceptor, whereas its two products are (S)-1-phenylethanol and reduced acceptor. This enzyme belongs to the family of oxidoreductases, specifically those acting on CH or CH2 groups with other acceptors. The systematic name of this enzyme class is ethylbenzene:acceptor oxidoreductase. Other names in common use include ethylbenzene dehydrogenase, and ethylbenzene:(acceptor) oxidoreductase. This enzyme participates in ethylbenzene degradation by ''Aromatoleum aromaticum'', a denitrifying bacterium related to the genera ''Azoarcus'' and ''Thauera''. It is a molybdenum enzyme belonging to the DMSO reductase family. Molybdenum enzymes are distinguished by the presence of a unique active site containing molybdenum atom, one or two molybdopte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzymology
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oxidoreductase
In biochemistry, an oxidoreductase is an enzyme that catalyzes the transfer of electrons from one molecule, the reductant, also called the electron donor, to another, the oxidant, also called the electron acceptor. This group of enzymes usually utilizes NADP+ or NAD+ as cofactors. Transmembrane oxidoreductases create electron transport chains in bacteria, chloroplasts and mitochondria, including respiratory complexes I, II and III. Some others can associate with biological membranes as peripheral membrane proteins or be anchored to the membranes through a single transmembrane helix. in ...
|
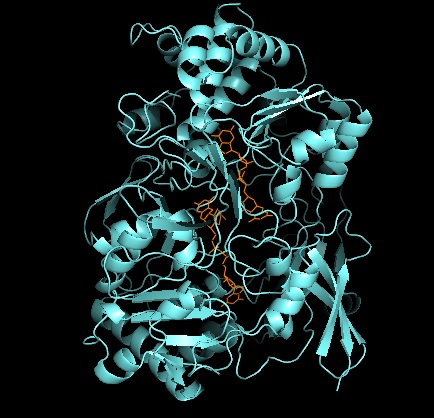
2ivf
In vitro fertilisation (IVF) is a process of fertilisation where an egg is combined with sperm in vitro ("in glass"). The process involves monitoring and stimulating an individual's ovulatory process, removing an ovum or ova (egg or eggs) from their ovaries and letting sperm fertilise them in a culture medium in a laboratory. After the fertilised egg (zygote) undergoes embryo culture for 2–6 days, it is transferred by catheter into the uterus, with the intention of establishing a successful pregnancy. IVF is a type of assisted reproductive technology used for infertility treatment, gestational surrogacy, and, in combination with pre-implantation genetic testing, avoiding transmission of genetic conditions. A fertilised egg from a donor may implant into a surrogate's uterus, and the resulting child is genetically unrelated to the surrogate. Some countries have banned or otherwise regulate the availability of IVF treatment, giving rise to fertility tourism. Restrictions ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules, such as proteins and nucleic acids. The data, typically obtained by X-ray crystallography, NMR spectroscopy, or, increasingly, cryo-electron microscopy, and submitted by biologists and biochemists from around the world, are freely accessible on the Internet via the websites of its member organisations (PDBe, PDBj, RCSB, and BMRB). The PDB is overseen by an organization called the Worldwide Protein Data Bank, wwPDB. The PDB is a key in areas of structural biology, such as structural genomics. Most major scientific journals and some funding agencies now require scientists to submit their structure data to the PDB. Many other databases use protein structures deposited in the PDB. For example, SCOP and CATH classify protein structures, while PDBsum provides a graphic overview of PDB entries using information from other sources, such as Gene ontology. History Two force ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tertiary Structure
Protein tertiary structure is the three dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains may interact and bond in a number of ways. The interactions and bonds of side chains within a particular protein determine its tertiary structure. The protein tertiary structure is defined by its atomic coordinates. These coordinates may refer either to a protein domain or to the entire tertiary structure.Branden C. and Tooze J. "Introduction to Protein Structure" Garland Publishing, New York. 1990 and 1991. A number of tertiary structures may fold into a quaternary structure.Kyte, J. "Structure in Protein Chemistry." Garland Publishing, New York. 1995. History The science of the tertiary structure of proteins has progressed from one of hypothesis to one of detailed definition. Although Emil Fischer had suggested proteins were made of polypept ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molybdopterin
Molybdopterins are a class of cofactors found in most molybdenum-containing and all tungsten-containing enzymes. Synonyms for molybdopterin are: MPT and pyranopterin-dithiolate. The nomenclature for this biomolecule can be confusing: Molybdopterin itself contains no molybdenum; rather, this is the name of the ligand (a ''pterin'') that will bind the active metal. After molybdopterin is eventually complexed with molybdenum, the complete ligand is usually called molybdenum cofactor. Molybdopterin consists of a pyranopterin, a complex heterocycle featuring a pyran fused to a pterin ring. In addition, the pyran ring features two thiolates, which serve as ligands in molybdo- and tungstoenzymes. In some cases, the alkyl phosphate group is replaced by an alkyl diphosphate nucleotide. Enzymes that contain the molybdopterin cofactor include xanthine oxidase, DMSO reductase, sulfite oxidase, and nitrate reductase. The only molybdenum-containing enzymes that do not feature molybdopteri ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DMSO Reductase
DMSO reductase is a molybdenum-containing enzyme that catalyzes reduction of dimethyl sulfoxide (DMSO) to dimethyl sulfide (DMS). This enzyme serves as the terminal reductase under anaerobic conditions in some bacteria, with DMSO being the terminal electron acceptor. During the course of the reaction, the oxygen atom in DMSO is transferred to molybdenum, and then reduced to water. DMSO reductase (DMSOR) and other members of the DMSO reductase family are unique to bacteria and archaea. Enzymes of this family in anaerobic oxidative phosphorylation and inorganic-donor-based lithotrophic respiration. These enzymes have been engineered to degrade oxoanions. DMSOR catalyzes the transfer of two electrons and one oxygen atom in the reaction: The active site of DMSOR contains molybdenum, which is otherwise rare in biology. Tertiary structure and active site As for other members of DMSO reductase family, the tertiary structure of DMSOR is composed of Mo-surrounding domains I-I ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ethylbenzene Degradation
Ethylbenzene is an organic compound with the formula . It is a highly flammable, colorless liquid with an odor similar to that of gasoline. This monocyclic aromatic hydrocarbon is important in the petrochemical industry as an reaction intermediate in the production of styrene, the precursor to polystyrene, a common plastic material. In 2012, more than 99% of ethylbenzene produced was consumed in the production of styrene. Occurrence and applications Ethylbenzene occurs naturally in coal tar and petroleum. The dominant application of ethylbenzene is its role as an intermediate in the production of polystyrene. Catalytic dehydrogenation of ethylbenzene gives hydrogen and styrene: : → C6H5CH=CH2 + As of May 2012, more than 99% of all the ethylbenzene produced is used for this purpose. Ethylbenzene hydroperoxide, a reagent and radical initiator, is produced by autoxidation of ethylbenzene: :C6H5CH2CH3 + O2 → C6H5CH(O2H)CH3 Niche uses Ethylbenzene is added t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Enzymes
This article lists enzymes by their classification in the International Union of Biochemistry and Molecular Biology's Enzyme Commission (EC) numbering system. * List of EC numbers (EC 5) * List of EC numbers (EC 6) :Oxidoreductases (EC 1) (Oxidoreductase) *Dehydrogenase * Luciferase *DMSO reductase :EC 1.1 (act on the CH-OH group of donors) * :EC 1.1.1 (with NAD+ or NADP+ as acceptor) ** Alcohol dehydrogenase (NAD) ** Alcohol dehydrogenase (NADP) **Homoserine dehydrogenase ** Aminopropanol oxidoreductase **Diacetyl reductase **Glycerol dehydrogenase **Propanediol-phosphate dehydrogenase ** glycerol-3-phosphate dehydrogenase (NAD+) ** D-xylulose reductase **L-xylulose reductase **Lactate dehydrogenase **Malate dehydrogenase **Isocitrate dehydrogenase ** HMG-CoA reductase * :EC 1.1.2 (with a cytochrome as acceptor) * :EC 1.1.3 (with oxygen as acceptor) **Glucose oxidase **L-gulonolactone oxidase **Thiamine oxidase **Xanthine oxidase * :EC 1.1.4 (with a disul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reduced Acceptor
An electron acceptor is a chemical entity that accepts electrons transferred to it from another compound. It is an oxidizing agent that, by virtue of its accepting electrons, is itself reduced in the process. Electron acceptors are sometimes mistakenly called electron receptors. Typical oxidizing agents undergo permanent chemical alteration through covalent or ionic reaction chemistry, resulting in the complete and irreversible transfer of one or more electrons. In many chemical circumstances, however, the transfer of electronic charge from an electron donor may be only fractional, meaning an electron is not completely transferred, but results in an electron resonance between the donor and acceptor. This leads to the formation of charge transfer complexes in which the components largely retain their chemical identities. The electron accepting power of an acceptor molecule is measured by its electron affinity which is the energy released when filling the lowest unoccupied molecul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |