|
DnaC
dnaC is a loading factor that complexes with the C-terminus of helicase dnaB and inhibits it from unwinding the dsDNA at a replication fork. A dnaB and dnaC associate near the dnaA bound origin for each of the ssDNA. One dnaB-dnaC complex is oriented in the opposite direction to the other dnaB-dnaC complex due to the antiparallel nature of DNA. Because they are oriented in opposite directions, one dnaB-dnaC complex will complex with dnaA from the N-terminus of dnaB whereas the other dnaB-dnaC complex will complex with dnaA from the dnaC. After the assembly of dnaG onto the N-terminus of dnaB, dnaC is released and dnaB will be allowed to begin unwinding dsDNA to make room for DNA polymerase III DNA polymerase III holoenzyme is the primary enzyme complex involved in prokaryotic DNA replication. It was discovered by Thomas Kornberg (son of Arthur Kornberg) and Malcolm Gefter in 1970. The complex has high processivity (i.e. the number of ... to begin synthesizing the daughter str ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DnaB Helicase
DnaB helicase is an enzyme in bacteria which opens the replication fork during DNA replication. Although the mechanism by which DnaB both couples ATP hydrolysis to translocation along DNA and denatures the duplex is unknown, a change in the quaternary structure of the protein involving dimerisation of the N-terminal domain has been observed and may occur during the enzymatic cycle. Initially when DnaB binds to dnaA, it is associated with dnaC, a negative regulator. After DnaC dissociates, DnaB binds dnaG. The N-terminal has a multi-helical structure that forms an orthogonal bundle. The C-terminal domain contains an ATP-binding site and is therefore probably the site of ATP hydrolysis. In eukaryotes, helicase function is provided by the MCM (Minichromosome maintenance) complex. The DnaB helicase is the product of the ''dnaB'' gene. The helicase enzyme that is produced is a hexamer in ''E. coli'', as well as in many other bacteria. The energy for DnaB activity is provided by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
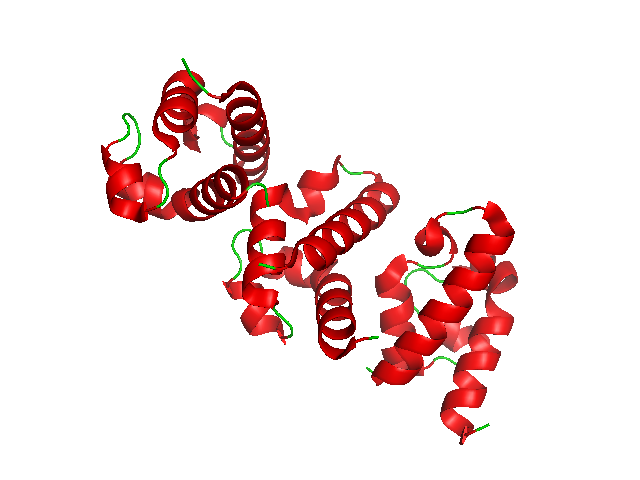
DnaA
Introduction Based on the Replicon Model, a positively active initiator molecule contacts with a particular spot on a circular chromosome called the replicator to start DNA replication. DnaA is a protein that activates initiation of DNA replication in bacteria. It is a replication initiation factor which promotes the unwinding of DNA at oriC. The DnaA proteins found in all bacteria engage with the DnaA boxes to start chromosomal replication. In addition to the DnaA protein, its concentration, binding to DnaA-boxes, and binding of ATP or ADP, we will cover the regulation of the DnaA gene, the unique characteristics of the DnaA gene expression, promoter strength, and translation efficiency. The onset of the initiation phase of DNA replication is determined by the concentration of DnaA. DnaA accumulates during growth and then triggers the initiation of replication. Replication begins with active DnaA binding to 9-mer (9-bp) repeats upstream of oriC. Binding of DnaA leads to strand s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Escherichia Coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus '' Escherichia'' that is commonly found in the lower intestine of warm-blooded organisms. Most ''E. coli'' strains are harmless, but some serotypes ( EPEC, ETEC etc.) can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains do not cause disease in humans and are part of the normal microbiota of the gut; such strains are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of ''E. coli'' benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between ''E ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Helicase
Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two hybridized nucleic acid strands (hence '' helic- + -ase''), using energy from ATP hydrolysis. There are many helicases, representing the great variety of processes in which strand separation must be catalyzed. Approximately 1% of eukaryotic genes code for helicases. The human genome codes for 95 non-redundant helicases: 64 RNA helicases and 31 DNA helicases. Many cellular processes, such as DNA replication, transcription, translation, recombination, DNA repair, and ribosome biogenesis involve the separation of nucleic acid strands that necessitates the use of helicases. Some specialized helicases are also involved in sensing of viral nucleic acids during infection and fulfill a immunological function. Function Helicases are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Polymerase III
DNA polymerase III holoenzyme is the primary enzyme complex involved in prokaryotic DNA replication. It was discovered by Thomas Kornberg (son of Arthur Kornberg) and Malcolm Gefter in 1970. The complex has high processivity (i.e. the number of nucleotides added per binding event) and, specifically referring to the replication of the '' E.coli'' genome, works in conjunction with four other DNA polymerases (Pol I, Pol II, Pol IV, and Pol V). Being the primary holoenzyme involved in replication activity, the DNA Pol III holoenzyme also has proofreading capabilities that corrects replication mistakes by means of exonuclease activity reading 3'→5' and synthesizing 5'→3'. DNA Pol III is a component of the replisome, which is located at the replication fork. Components The replisome is composed of the following: *2 DNA Pol III enzymes, each comprising α, ε and θ subunits. (It has been proven that there is a third copy of Pol III at the replisome.) **the α subunit (encod ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenosine Triphosphate
Adenosine triphosphate (ATP) is an organic compound that provides energy to drive many processes in living cells, such as muscle contraction, nerve impulse propagation, condensate dissolution, and chemical synthesis. Found in all known forms of life, ATP is often referred to as the "molecular unit of currency" of intracellular energy transfer. When consumed in metabolic processes, it converts either to adenosine diphosphate (ADP) or to adenosine monophosphate (AMP). Other processes regenerate ATP. The human body recycles its own body weight equivalent in ATP each day. It is also a precursor to DNA and RNA, and is used as a coenzyme. From the perspective of biochemistry, ATP is classified as a nucleoside triphosphate, which indicates that it consists of three components: a nitrogenous base ( adenine), the sugar ribose, and the triphosphate. Structure ATP consists of an adenine attached by the 9-nitrogen atom to the 1′ carbon atom of a sugar ( ribose), which in tu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacterial Proteins
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic relationships ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |


.jpg)
