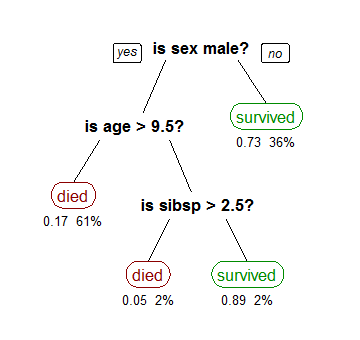|
Discovery Studio
Discovery Studio is a suite of software for simulating small molecule and macromolecule systems. It is developed and distributed bDassault Systemes BIOVIA(formerly Accelrys). The product suite has a stronacademic collaboration programme supporting scientific research and makes use of a number of software algorithms developed originally in the scientific community, including CHARMM, MODELLER, DELPHI, ZDOCK, DMol3 and more. Scope Discovery Studio provides software applications covering the following areas: * Simulations ** Including Molecular Mechanics, Molecular Dynamics, Quantum Mechanics ** For molecular mechanics based simulations: Include implicit and explicit-based solvent models and membrane models ** Also includes the ability to perform hybrid QM/MM calculations * Ligand Design ** Including tools for enumerating molecular libraries and library optimization * Pharmacophore modeling ** Including creation, validation and virtual screening * Structure-based Design ** Inclu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Software
Software is a set of computer programs and associated documentation and data. This is in contrast to hardware, from which the system is built and which actually performs the work. At the lowest programming level, executable code consists of machine language instructions supported by an individual processor—typically a central processing unit (CPU) or a graphics processing unit (GPU). Machine language consists of groups of binary values signifying processor instructions that change the state of the computer from its preceding state. For example, an instruction may change the value stored in a particular storage location in the computer—an effect that is not directly observable to the user. An instruction may also invoke one of many input or output operations, for example displaying some text on a computer screen; causing state changes which should be visible to the user. The processor executes the instructions in the order they are provided, unless it is instructed ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein-protein Docking
Macromolecular docking is the computational modelling of the quaternary structure of complexes formed by two or more interacting biological macromolecules. Protein–protein complexes are the most commonly attempted targets of such modelling, followed by protein–nucleic acid complexes. The ultimate goal of docking is the prediction of the three-dimensional structure of the macromolecular complex of interest as it would occur in a living organism. Docking itself only produces plausible candidate structures. These candidates must be ranked using methods such as scoring functions to identify structures that are most likely to occur in nature. The term "docking" originated in the late 1970s, with a more restricted meaning; then, "docking" meant refining a model of a complex structure by optimizing the separation between the interactors but keeping their relative orientations fixed. Later, the relative orientations of the interacting partners in the modelling was allowed to vary, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homology Modelling
Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the "''target''" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the "''template''"). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been seen that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure. Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure. It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Design Software
Molecular design software is notable software for molecular modeling, that provides special support for developing molecular models ''de novo''. In contrast to the usual molecular modeling programs, such as for molecular dynamics and quantum chemistry, such software ''directly'' supports the aspects related to constructing molecular models, including: * Molecular graphics * interactive molecular drawing and conformational editing * building polymeric molecules, crystals, and solvated systems * partial charges development * geometry optimization * support for the different aspects of force field development Comparison of software covering the major aspects of molecular design Notes and references See also {{columns-list, colwidth=30em, *Molecule editor *Molecular modelling *Molecular modeling on GPUs *Protein design *Drug design *Force field (chemistry) *Comparison of force field implementations *Comparison of nucleic acid simulation software *Comparison of software for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Modelling
Molecular modelling encompasses all methods, theoretical and computational, used to model or mimic the behaviour of molecules. The methods are used in the fields of computational chemistry, drug design, computational biology and materials science to study molecular systems ranging from small chemical systems to large biological molecules and material assemblies. The simplest calculations can be performed by hand, but inevitably computers are required to perform molecular modelling of any reasonably sized system. The common feature of molecular modelling methods is the atomistic level description of the molecular systems. This may include treating atoms as the smallest individual unit (a molecular mechanics approach), or explicitly modelling protons and neutrons with its quarks, anti-quarks and gluons and electrons with its photons (a quantum chemistry approach). Molecular mechanics Molecular mechanics is one aspect of molecular modelling, as it involves the use of classical ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Quantum Chemistry Computer Programs
Quantum chemistry computer programs are used in computational chemistry to implement the methods of quantum chemistry. Most include the Hartree–Fock (HF) and some post-Hartree–Fock methods. They may also include density functional theory (DFT), molecular mechanics or semi-empirical quantum chemistry methods. The programs include both open source Open source is source code that is made freely available for possible modification and redistribution. Products include permission to use the source code, design documents, or content of the product. The open-source model is a decentralized sof ... and commercial software. Most of them are large, often containing several separate programs, and have been developed over many years. Overview The following tables illustrates some of the main capabilities of notable packages: Numerical details Quantum chemistry and solid-state physics characteristics Post processing packages in quantum chemistry and solid-state physics ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Software For Molecular Mechanics Modeling
This is a list of computer programs that are predominantly used for molecular mechanics calculations. See also *Car–Parrinello molecular dynamics *Comparison of force-field implementations *Comparison of nucleic acid simulation software *List of molecular graphics systems *List of protein structure prediction software *List of quantum chemistry and solid-state physics software *List of software for Monte Carlo molecular modeling *List of software for nanostructures modeling *Molecular design software *Molecular dynamics *Molecular modeling on GPUs *Molecule editor A molecule editor is a computer program for creating and modifying representations of chemical structures. Molecule editors can manipulate chemical structure representations in either a simulated two-dimensional space or three-dimensional space, v ... Notes and references External linksSINCRIS [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ADME
ADME is an abbreviation in pharmacokinetics and pharmacology for " absorption, distribution, metabolism, and excretion", and describes the disposition of a pharmaceutical compound within an organism. The four criteria all influence the drug levels and kinetics of drug exposure to the tissues and hence influence the performance and pharmacological activity of the compound as a drug. Sometimes, liberation and/or toxicity are also considered, yielding LADME, ADMET, or LADMET. Components Absorption/administration For a compound to reach a tissue, it usually must be taken into the bloodstream – often via mucous surfaces like the digestive tract (intestinal absorption) – before being taken up by the target cells. Factors such as poor compound solubility, gastric emptying time, intestinal transit time, chemical instability in the stomach, and inability to permeate the intestinal wall can all reduce the extent to which a drug is absorbed after oral administration. Absorptio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Recursive Partitioning
Recursive partitioning is a statistical method for multivariable analysis. Recursive partitioning creates a decision tree that strives to correctly classify members of the population by splitting it into sub-populations based on several dichotomous independent variables. The process is termed recursive because each sub-population may in turn be split an indefinite number of times until the splitting process terminates after a particular stopping criterion is reached. Recursive partitioning methods have been developed since the 1980s. Well known methods of recursive partitioning include Ross Quinlan's ID3 algorithm and its successors, C4.5 and C5.0 and Classification and Regression Trees (CART). Ensemble learning methods such as Random Forests help to overcome a common criticism of these methods – their vulnerability to overfitting of the data – by employing different algorithms and combining their output in some way. This article focuses on recursive partitioning for medic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Partial Least Squares
Partial least squares regression (PLS regression) is a statistical method that bears some relation to principal components regression; instead of finding hyperplanes of maximum variance between the response and independent variables, it finds a linear regression model by projecting the predicted variables and the observable variables to a new space. Because both the ''X'' and ''Y'' data are projected to new spaces, the PLS family of methods are known as bilinear factor models. Partial least squares discriminant analysis (PLS-DA) is a variant used when the Y is categorical. PLS is used to find the fundamental relations between two matrices (''X'' and ''Y''), i.e. a latent variable approach to modeling the covariance structures in these two spaces. A PLS model will try to find the multidimensional direction in the ''X'' space that explains the maximum multidimensional variance direction in the ''Y'' space. PLS regression is particularly suited when the matrix of predictors has more ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Multiple Linear Regression
In statistics, linear regression is a linear approach for modelling the relationship between a scalar response and one or more explanatory variables (also known as dependent and independent variables). The case of one explanatory variable is called ''simple linear regression''; for more than one, the process is called multiple linear regression. This term is distinct from multivariate linear regression, where multiple correlated dependent variables are predicted, rather than a single scalar variable. In linear regression, the relationships are modeled using linear predictor functions whose unknown model parameters are estimated from the data. Such models are called linear models. Most commonly, the conditional mean of the response given the values of the explanatory variables (or predictors) is assumed to be an affine function of those values; less commonly, the conditional median or some other quantile is used. Like all forms of regression analysis, linear regression focuses on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |