|
DEC2
"Basic helix-loop-helix family, member e41", or BHLHE41, is a gene that encodes a basic helix-loop-helix transcription factor repressor protein in various tissues of both humans and mice. It is also known as DEC2, hDEC2, and SHARP1, and was previously known as "basic helix-loop-helix domain containing, class B, 3", or BHLHB3. BHLHE41 is known for its role in the circadian molecular mechanisms that influence sleep quantity as well as its role in immune function and the maturation of T helper type 2 cell lineages associated with humoral immunity. History DrKlaus-Armin Naves lab identified BHLHE41/SHARP1 and BHLHE40/SHARP2 as a novel subfamily in the basic helix-loop-helix (BHLH) protein family. They differentiated BHLHE41/SHARP1 and BHLHE40/SHARP2 from other BHLH-protein encoding genes since they are not transcribed until the end of embryonic development. The DNA sequence of BHLHE41 was first obtained by Dr. Yukia Kato's lab through a cDNA library search. Particularly, t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BHLHB2
Class E basic helix-loop-helix protein 40 is a protein that in humans is encoded by the ''BHLHE40'' gene In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a ba .... Function DEC1 encodes a basic helix-loop-helix protein expressed in various tissues. Expression in the chondrocytes is responsive to the addition of Bt2cAMP. Differentiated embryo chondrocyte expressed gene 1 is believed to be involved in the control of cell differentiation. References Further reading * * * * * * * * * * * * * * * * External links * * * * * Transcription factors {{gene-3-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Base Pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, "Watson–Crick" (or "Watson–Crick–Franklin") base pairs (guanine–cytosine and adenine–thymine) allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The Complementarity (molecular biology), complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Circadian Clock
A circadian clock, or circadian oscillator, is a biochemical oscillator that cycles with a stable phase (waves), phase and is synchronized with solar time. Such a clock's ''in vivo'' period is necessarily almost exactly 24 hours (the earth's current day, solar day). In most living things, internally synchronized circadian clocks make it possible for the organism to anticipate daily environmental changes corresponding with the day–night cycle and adjust its biology and behavior accordingly. The term circadian derives from the Latin ''circa'' (about) ''dies'' (a day), since when taken away from external cues (such as environmental light), they do not run to exactly 24 hours. Clocks in humans in a lab in constant low light, for example, will average about 24.2 hours per day, rather than 24 hours exactly. The normal body clock oscillates with an endogenous period of exactly 24 hours, it Entrainment (chronobiology), entrains, when it receives sufficient daily corrective signals from ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Regulator Gene
A regulator gene, regulator, or regulatory gene is a gene involved in controlling the expression of one or more other genes. Regulatory sequences, which encode regulatory genes, are often at the five prime end (5') to the start site of transcription of the gene they regulate. In addition, these sequences can also be found at the three prime end (3') to the transcription start site. In both cases, whether the regulatory sequence occurs before (5') or after (3') the gene it regulates, the sequence is often many kilobases away from the transcription start site. A regulator gene may encode a protein, or it may work at the level of RNA, as in the case of genes encoding microRNAs. An example of a regulator gene is a gene that codes for a repressor protein that inhibits the activity of an operator (a gene which binds repressor proteins thus inhibiting the translation of RNA to protein via RNA polymerase). In prokaryotes, regulator genes often code for repressor proteins. Repressor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Repressor
In molecular genetics, a repressor is a DNA- or RNA-binding protein that inhibits the expression of one or more genes by binding to the operator or associated silencers. A DNA-binding repressor blocks the attachment of RNA polymerase to the promoter, thus preventing transcription of the genes into messenger RNA. An RNA-binding repressor binds to the mRNA and prevents translation of the mRNA into protein. This blocking or reducing of expression is called repression. Function If an inducer, a molecule that initiates the gene expression, is present, then it can interact with the repressor protein and detach it from the operator. RNA polymerase then can transcribe the message (expressing the gene). A co-repressor is a molecule that can bind to the repressor and make it bind to the operator tightly, which decreases transcription. A repressor that binds with a co-repressor is termed an ''aporepressor'' or ''inactive repressor''. One type of aporepressor is the trp repressor, a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Suprachiasmatic Nucleus
The suprachiasmatic nucleus or nuclei (SCN) is a tiny region of the brain in the hypothalamus, situated directly above the optic chiasm. It is responsible for controlling circadian rhythms. The neuronal and hormonal activities it generates regulate many different body functions in a 24-hour cycle. The mouse SCN contains approximately 20,000 neurons. The SCN interacts with many other regions of the brain. It contains several cell types and several different peptides (including vasopressin and vasoactive intestinal peptide) and neurotransmitters. Neuroanatomy The SCN is situated in the anterior part of the hypothalamus immediately dorsal, or ''superior'' (hence supra) to the optic chiasm (CHO) bilateral to (on either side of) the third ventricle. The nucleus can be divided into ventrolateral and dorsolateral portions, also known as the core and shell, respectively. These regions differ in their expression of the clock genes, the core expresses them in response to stimuli whereas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EHMT2
Euchromatic histone-lysine N-methyltransferase 2 (EHMT2), also known as G9a, is a histone methyltransferase enzyme that in humans is encoded by the ''EHMT2'' gene. G9a catalyzes the mono- and di-methylated states of histone H3 at lysine residue 9 (i.e., H3K9me1 and H3K9me2) and lysine residue 27 ( H3K27me1 and HeK27me2). Function A cluster of genes, BAT1-BAT5, has been localized in the vicinity of the genes for TNF alpha and TNF beta. This gene is found near this cluster; it was mapped near the gene for C2 within a 120-kb region that included a HSP70 gene pair. These genes are all within the human major histocompatibility complex class III region. This gene was thought to be two different genes, NG36 and G9a, adjacent to each other but a recent publication shows that there is only a single gene. The protein encoded by this gene is thought to be involved in intracellular protein-protein interaction. There are three alternatively spliced transcript variants of this gene but onl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone Methyltransferase
Histone methyltransferases (HMT) are histone-modifying enzymes (e.g., histone-lysine N-methyltransferases and histone-arginine N-methyltransferases), that catalyze the transfer of one, two, or three methyl groups to lysine and arginine residues of histone proteins. The attachment of methyl groups occurs predominantly at specific lysine or arginine residues on histones H3 and H4. Two major types of histone methyltranferases exist, lysine-specific (which can be SET (Su(var)3-9, Enhancer of Zeste, Trithorax) domain containing or non-SET domain containing) and arginine-specific. In both types of histone methyltransferases, S-Adenosyl methionine (SAM) serves as a cofactor and methyl donor group. The genomic DNA of eukaryotes associates with histones to form chromatin. The level of chromatin compaction depends heavily on histone methylation and other post-translational modifications of histones. Histone methylation is a principal epigenetic modification of chromatin that determines ge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-terminus
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus. Chemistry Each amino acid has a carboxyl group and an amine group. Amino acids link to one another to form a chain by a dehydration reaction which joins the amine group of one amino acid to the carboxyl group of the next. Thus polypeptide chains have an end with an unbound carboxyl group, the C-terminus, and an end with an unbound amine group, the N-terminus. Proteins are naturally synthesized starting from the N-terminus and ending at the C-terminus. Function C-terminal retention signals While the N-terminus of a protein often c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glycine
Glycine (symbol Gly or G; ) is an amino acid that has a single hydrogen atom as its side chain. It is the simplest stable amino acid (carbamic acid is unstable), with the chemical formula NH2‐ CH2‐ COOH. Glycine is one of the proteinogenic amino acids. It is encoded by all the codons starting with GG (GGU, GGC, GGA, GGG). Glycine is integral to the formation of alpha-helices in secondary protein structure due to its compact form. For the same reason, it is the most abundant amino acid in collagen triple-helices. Glycine is also an inhibitory neurotransmitter – interference with its release within the spinal cord (such as during a ''Clostridium tetani'' infection) can cause spastic paralysis due to uninhibited muscle contraction. It is the only achiral proteinogenic amino acid. It can fit into hydrophilic or hydrophobic environments, due to its minimal side chain of only one hydrogen atom. History and etymology Glycine was discovered in 1820 by the French chemist He ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
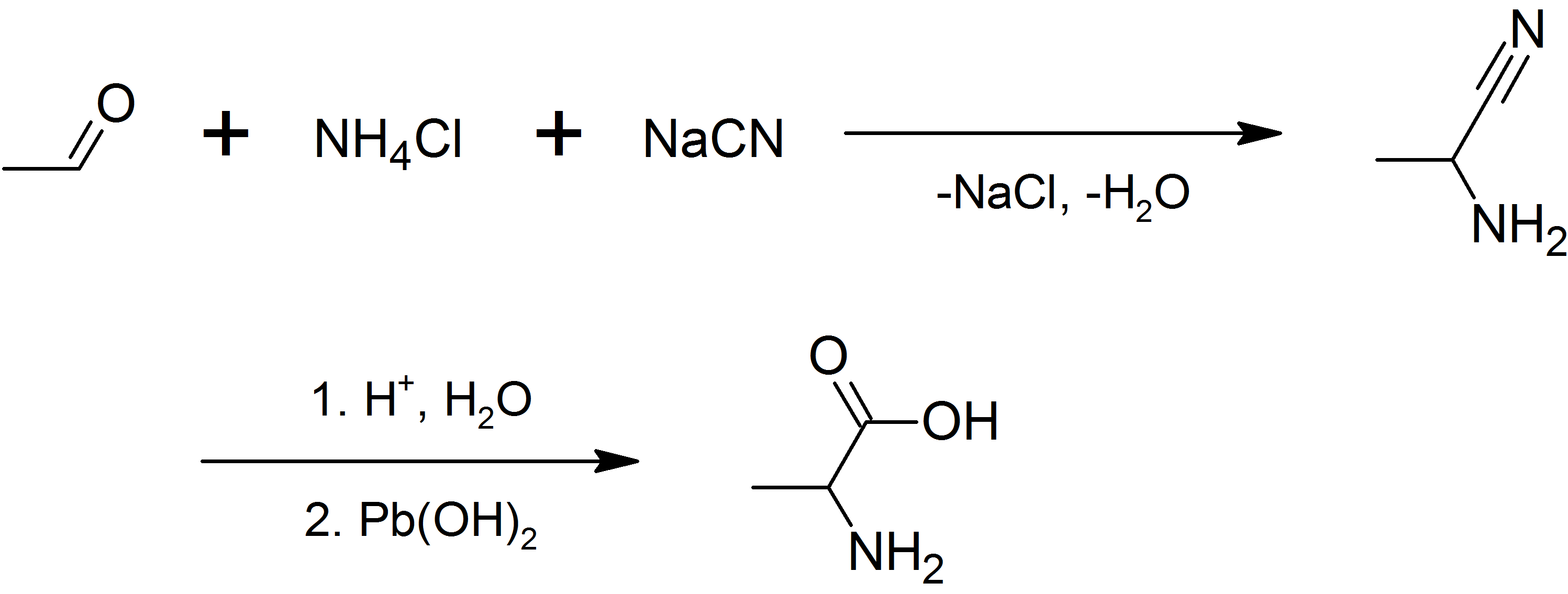
Alanine
Alanine (symbol Ala or A), or α-alanine, is an α-amino acid that is used in the biosynthesis of proteins. It contains an amine group and a carboxylic acid group, both attached to the central carbon atom which also carries a methyl group side chain. Consequently, its IUPAC systematic name is 2-aminopropanoic acid, and it is classified as a nonpolar, aliphatic α-amino acid. Under biological conditions, it exists in its zwitterionic form with its amine group protonated (as −NH3+) and its carboxyl group deprotonated (as −CO2−). It is non-essential to humans as it can be synthesised metabolically and does not need to be present in the diet. It is encoded by all codons starting with GC (GCU, GCC, GCA, and GCG). The L-isomer of alanine (left-handed) is the one that is incorporated into proteins. L-alanine is second only to leucine in rate of occurrence, accounting for 7.8% of the primary structure in a sample of 1,150 proteins. The right-handed form, D-alanine, occurs in p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |