|
ConsensusPathDB
The ConsensusPathDB is a molecular functional interaction database, integrating information on protein interactions, genetic interactions signaling, metabolism, gene regulation, and drug-target interactions in humans. ConsensusPathDB currently (release 30) includes such interactions from 32 databases. ConsensusPathDB is freely available for academic use under http://ConsensusPathDB.org. Integrated Databases * Reactome (Metabolic pathway, metabolic and signaling pathways) * KEGG (metabolic pathways only have been integrated in ConsensusPathDB) * HumanCyc (metabolic pathways) * PID - Pathway Interaction Database (signaling pathways) * BioCarta (signaling pathways) * Netpath (signaling pathways) * IntAct (protein interactions) * Database of Interacting Proteins, DIP (protein interactions) * MINT (protein interactions) * Human Protein Reference Database, HPRD (protein interactions) * BioGRID (protein interactions) * SPIKE (protein interactions, signaling reactions) * WikiPathways (Metab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BioPAX
BioPAX (Biological Pathway Exchange) is a RDF/OWL-based standard language to represent biological pathways at the molecular and cellular level. Its major use is to facilitate the exchange of pathway data. Pathway data captures our understanding of biological processes, but its rapid growth necessitates development of databases and computational tools to aid interpretation. However, the current fragmentation of pathway information across many databases with incompatible formats presents barriers to its effective use. BioPAX solves this problem by making pathway data substantially easier to collect, index, interpret and share. BioPAX can represent metabolic and signaling pathways, molecular and genetic interactions and gene regulation networks. BioPAX was created through a community process. Through BioPAX, millions of interactions organized into thousands of pathways across many organisms, from a growing number of sources, are available. Thus, large amounts of pathway data are availa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biochemical Pathway
In biochemistry, a metabolic pathway is a linked series of chemical reactions occurring within a cell. The reactants, products, and intermediates of an enzymatic reaction are known as metabolites, which are modified by a sequence of chemical reactions catalyzed by enzymes. In most cases of a metabolic pathway, the product of one enzyme acts as the substrate for the next. However, side products are considered waste and removed from the cell. Different metabolic pathways function in the position within a eukaryotic cell and the significance of the pathway in the given compartment of the cell. For instance, the electron transport chain and oxidative phosphorylation all take place in the mitochondrial membrane. In contrast, glycolysis, pentose phosphate pathway, and fatty acid biosynthesis all occur in the cytosol of a cell. There are two types of metabolic pathways that are characterized by their ability to either synthesize molecules with the utilization of energy ( anab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metabolic Pathway
In biochemistry, a metabolic pathway is a linked series of chemical reactions occurring within a cell (biology), cell. The reactants, products, and Metabolic intermediate, intermediates of an enzymatic reaction are known as metabolites, which are modified by a sequence of chemical reactions catalyzed by enzymes. In most cases of a metabolic pathway, the product (chemistry), product of one enzyme acts as the substrate (chemistry), substrate for the next. However, side products are considered waste and removed from the cell. Different metabolic pathways function in the position within a Eukaryotic Cell, eukaryotic cell and the significance of the pathway in the given compartment of the cell. For instance, the electron transport chain and oxidative phosphorylation all take place in the mitochondrial membrane. In contrast, glycolysis, pentose phosphate pathway, and Fatty acid synthesis, fatty acid biosynthesis all occur in the cytosol of a cell. There are two types of metabolic pathw ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
KEGG
KEGG (Kyoto Encyclopedia of Genes and Genomes) is a collection of databases dealing with genomes, biological pathways, diseases, drugs, and chemical substances. KEGG is utilized for bioinformatics research and education, including data analysis in genomics, metagenomics, metabolomics and other omics studies, modeling and simulation in systems biology, and translational research in drug development. The KEGG database project was initiated in 1995 by Minoru Kanehisa, professor at the Institute for Chemical Research, Kyoto University, under the then ongoing Japanese Human Genome Program. Foreseeing the need for a computerized resource that can be used for biological interpretation of genome sequence data, he started developing the KEGG PATHWAY database. It is a collection of manually drawn KEGG pathway maps representing experimental knowledge on metabolism and various other functions of the cell and the organism. Each pathway map contains a network of molecular interactions and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Database
In computing, a database is an organized collection of data or a type of data store based on the use of a database management system (DBMS), the software that interacts with end users, applications, and the database itself to capture and analyze the data. The DBMS additionally encompasses the core facilities provided to administer the database. The sum total of the database, the DBMS and the associated applications can be referred to as a database system. Often the term "database" is also used loosely to refer to any of the DBMS, the database system or an application associated with the database. Before digital storage and retrieval of data have become widespread, index cards were used for data storage in a wide range of applications and environments: in the home to record and store recipes, shopping lists, contact information and other organizational data; in business to record presentation notes, project research and notes, and contact information; in schools as flash c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
WikiPathways
WikiPathways is a community resource for contributing and maintaining content dedicated to biological pathways. Any registered WikiPathways user can contribute, and anybody can become a registered user. Contributions are monitored by a group of admins, but the bulk of peer review, editorial curation, and maintenance is the responsibility of the user community. WikiPathways is originally built using MediaWiki software, a custom graphical pathway editing tool ( PathVisio) and integrated BridgeDb databases covering major gene, protein, and metabolite systems. WikiPathways was founded in 2008 by Thomas Kelder, Alex Pico, Martijn Van Iersel, Kristina Hanspers, Bruce Conklin and Chris Evelo. Current architects are Alex Pico and Martina Summer-Kutmon. Pathway content Each article at WikiPathways is dedicated to a particular pathway. Many types of molecular pathways are covered, including metabolic, signaling, regulatory, etc. and the supported species include human, mouse, zebrafish, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
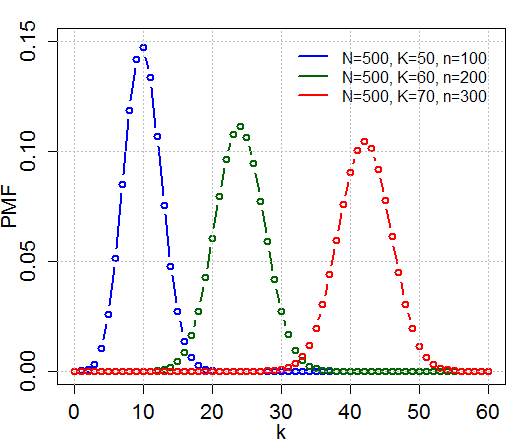
Hypergeometric Distribution
In probability theory and statistics, the hypergeometric distribution is a Probability distribution#Discrete probability distribution, discrete probability distribution that describes the probability of k successes (random draws for which the object drawn has a specified feature) in n draws, ''without'' replacement, from a finite Statistical population, population of size N that contains exactly K objects with that feature, wherein each draw is either a success or a failure. In contrast, the binomial distribution describes the probability of k successes in n draws ''with'' replacement. Definitions Probability mass function The following conditions characterize the hypergeometric distribution: * The result of each draw (the elements of the population being sampled) can be classified into one of Binary variable, two mutually exclusive categories (e.g. Pass/Fail or Employed/Unemployed). * The probability of a success changes on each draw, as each draw decreases the population ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
P-value
In null-hypothesis significance testing, the ''p''-value is the probability of obtaining test results at least as extreme as the result actually observed, under the assumption that the null hypothesis is correct. A very small ''p''-value means that such an extreme observed outcome would be very unlikely ''under the null hypothesis''. Even though reporting ''p''-values of statistical tests is common practice in academic publications of many quantitative fields, misinterpretation and misuse of p-values is widespread and has been a major topic in mathematics and metascience. In 2016, the American Statistical Association (ASA) made a formal statement that "''p''-values do not measure the probability that the studied hypothesis is true, or the probability that the data were produced by random chance alone" and that "a ''p''-value, or statistical significance, does not measure the size of an effect or the importance of a result" or "evidence regarding a model or hypothesis". That ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
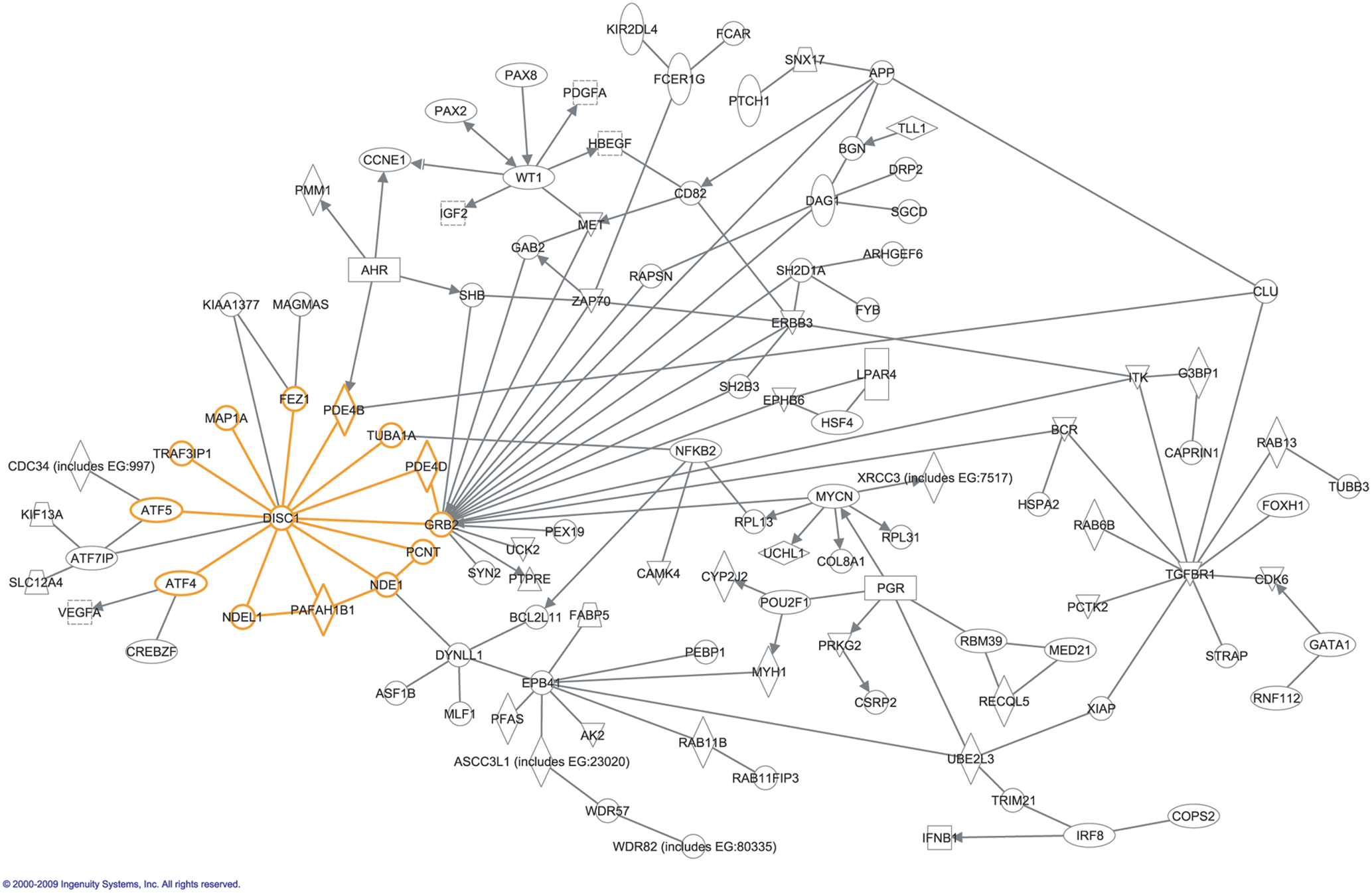
Interaction Network
In molecular biology, an interactome is the whole set of molecular interactions in a particular cell. The term specifically refers to physical interactions among molecules (such as those among proteins, also known as protein–protein interactions, PPIs; or between small molecules and proteins) but can also describe sets of indirect interactions among genes ( genetic interactions). The word "interactome" was originally coined in 1999 by a group of French scientists headed by Bernard Jacq. Mathematically, interactomes are generally displayed as graphs. While interactomes may be described as biological networks, they should not be confused with other networks such as neural networks or food webs. Molecular interaction networks Molecular interactions can occur between molecules belonging to different biochemical families (proteins, nucleic acids, lipids, carbohydrates, etc.) and also within a given family. Whenever such molecules are connected by physical interactions, they form molec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UniProt
UniProt is a freely accessible database of protein sequence and functional information, many entries being derived from genome sequencing projects. It contains a large amount of information about the biological function of proteins derived from the research literature. It is maintained by the UniProt consortium, which consists of several European bioinformatics organisations and a foundation from Washington, DC, USA. The UniProt consortium The UniProt consortium comprises the European Bioinformatics Institute (EBI), the Swiss Institute of Bioinformatics (SIB), and the Protein Information Resource (PIR). EBI, located at the Wellcome Trust Genome Campus in Hinxton, UK, hosts a large resource of bioinformatics databases and services. SIB, located in Geneva, Switzerland, maintains the ExPASy (Expert Protein Analysis System) servers that are a central resource for proteomics tools and databases. PIR, hosted by the National Biomedical Research Foundation (NBRF) at the George ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metabolite
In biochemistry, a metabolite is an intermediate or end product of metabolism. The term is usually used for small molecules. Metabolites have various functions, including fuel, structure, signaling, stimulatory and inhibitory effects on enzymes, catalytic activity of their own (usually as a cofactor to an enzyme), defense, and interactions with other organisms (e.g. pigments, odorants, and pheromones). A primary metabolite is directly involved in normal "growth", development, and reproduction. Ethylene exemplifies a primary metabolite produced large-scale by industrial microbiology. A secondary metabolite is not directly involved in those processes, but usually has an important ecological function. Examples include antibiotics and pigments such as resins and terpenes etc. Some antibiotics use primary metabolites as precursors, such as actinomycin, which is created from the primary metabolite tryptophan. Some sugars are metabolites, such as fructose or glucose, which ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |