|
Circular DNA (other)
{{disambiguation ...
Circular DNA is DNA that forms a closed loop and has no ends. Examples include: * Plasmids, mobile genetic elements * cccDNA, formed by some viruses inside cell nuclei * Circular bacterial chromosomes * Mitochondrial DNA (mtDNA) * Chloroplast DNA (cpDNA), and that of other plastids *Extrachromosomal circular DNA (eccDNA) See also * Inverse polymerase chain reaction (Inverse PCR), technique for finding unknown DNA flanking known DNA * Effect of circular DNA on gel electrophoresis * Cyclic nucleotide * Cyclic adenosine monophosphate (cAMP) * Cyclic guanosine monophosphate (cGMP) * Circular RNA * Cyclic peptide Cyclic peptides are polypeptide chains which contain a circular sequence of bonds. This can be through a connection between the amino and carboxyl ends of the peptide, for example in cyclosporin; a connection between the amino end and a side chai ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria; however, plasmids are sometimes present in archaea and eukaryotic organisms. In nature, plasmids often carry genes that benefit the survival of the organism and confer selective advantage such as antibiotic resistance. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain only additional genes that may be useful in certain situations or conditions. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the inter ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CccDNA
cccDNA (covalently closed circular DNA) is a special DNA structure that arises during the propagation of some viruses in the cell nucleus and may remain permanently there. It is a double-stranded DNA that originates in a linear form that is ligated by means of DNA ligase to a covalently closed ring. In most cases, transcription of viral DNA can occur from the circular form only. The cccDNA of viruses is also known as episomal DNA or occasionally as a minichromosome. cccDNA was first described in bacteriophages, but it was also found in some cell cultures where an infection of DNA viruses (''Polyomaviridae'') was detected. cccDNA is typical of ''Caulimoviridae'' and ''Hepadnaviridae'', including the hepatitis B virus (HBV). cccDNA in HBV is formed by conversion of capsid-associated relaxed circular DNA (rcDNA). Following hepatitis B infections, cccDNA can remain following clinical treatment in liver cells and can rarely reactivate. The relative quantity of cccDNA present is a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Circular Bacterial Chromosome
A circular chromosome is a chromosome in bacteria, archaea, mitochondria, and chloroplasts, in the form of a molecule of circular DNA, unlike the linear chromosome of most eukaryotes. Most prokaryote chromosomes contain a circular DNA molecule – there are no free ends to the DNA. Free ends would otherwise create significant challenges to cells with respect to DNA replication and stability. Cells that do contain chromosomes with DNA ends, or telomeres (most eukaryotes), have acquired elaborate mechanisms to overcome these challenges. However, a circular chromosome can provide other challenges for cells. After replication, the two progeny circular chromosomes can sometimes remain interlinked or tangled, and they must be resolved so that each cell inherits one complete copy of the chromosome during cell division. Replication The circular bacteria chromosome replication is best understood in the well-studied bacteria ''Escherichia coli'' and ''Bacillus subtilis''. Chromoso ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitochondrial DNA
Mitochondrial DNA (mtDNA or mDNA) is the DNA located in mitochondria, cellular organelles within eukaryotic cells that convert chemical energy from food into a form that cells can use, such as adenosine triphosphate (ATP). Mitochondrial DNA is only a small portion of the DNA in a eukaryotic cell; most of the DNA can be found in the cell nucleus and, in plants and algae, also in plastids such as chloroplasts. Human mitochondrial DNA was the first significant part of the human genome to be sequenced. This sequencing revealed that the human mtDNA includes 16,569 base pairs and encodes 13 proteins. Since animal mtDNA evolves faster than nuclear genetic markers, it represents a mainstay of phylogenetics and evolutionary biology. It also permits an examination of the relatedness of populations, and so has become important in anthropology and biogeography. Origin Nuclear and mitochondrial DNA are thought to be of separate evolutionary origin, with the mtDNA being derived ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chloroplast DNA
Chloroplast DNA (cpDNA) is the DNA located in chloroplasts, which are photosynthetic organelles located within the cells of some eukaryotic organisms. Chloroplasts, like other types of plastid, contain a genome separate from that in the cell nucleus. The existence of chloroplast DNA was identified biochemically in 1959, and confirmed by electron microscopy in 1962. The discoveries that the chloroplast contains ribosomes and performs protein synthesis revealed that the chloroplast is genetically semi-autonomous. The first complete chloroplast genome sequences were published in 1986, ''Nicotiana tabacum'' (tobacco) by Sugiura and colleagues and ''Marchantia polymorpha'' (liverwort) by Ozeki et al. Since then, a great number of chloroplast DNAs from various species have been sequenced. Molecular structure Chloroplast DNAs are circular, and are typically 120,000–170,000 base pairs long. They can have a contour length of around 30–60 micrometers, and have a mass of about 80� ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Extrachromosomal Circular DNA
Extrachromosomal circular DNA (eccDNA) is a type of double-stranded circular DNA structure that was first discovered in 1964 by Alix Bassel and Yasuo Hotta. In contrast to previously identified circular DNA structures (e.g., bacterial plasmids, mitochondrial DNA, circular bacterial chromosomes, or chloroplast DNA), eccDNA are circular DNA found in the eukaryotic nuclei of plant and animal (including human) cells. Extrachromosomal circular DNA is derived from chromosomal DNA, can range in size from 50 base pairs to several mega-base pairs in length, and can encode regulatory elements and full-length genes. eccDNA has been observed in various eukaryotic species and it is proposed to be a byproduct of programmed DNA recombination events, such as V(D)J recombination. Historical Background In 1964, Bassel and Hotta published their initial discovery of eccDNA that they made while researching Franklin Stahl’s chromosomal theory. In their experiments, they visualized isolated whea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Inverse Polymerase Chain Reaction
Inverse polymerase chain reaction (Inverse PCR) is a variant of the polymerase chain reaction that is used to amplify DNA with only one known sequence. One limitation of conventional PCR is that it requires primers complementary to both termini of the target DNA, but this method allows PCR to be carried out even if only one sequence is available from which primers may be designed. Inverse PCR is especially useful for the determination of insert locations. For example, various retroviruses and transposons randomly integrate into genomic DNA. To identify the sites where they have entered, the known, "internal" viral or transposon sequences can be used to design primers that will amplify a small portion of the flanking, "external" genomic DNA. The amplified product can then be sequenced and compared with DNA databases to locate the sequence which has been disrupted. The inverse PCR method involves a series of restriction digests and ligation, resulting in a looped fragment that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gel Electrophoresis Of Nucleic Acids
Nucleic acid electrophoresis is an analytical technique used to separate DNA or RNA fragments by size and reactivity. Nucleic acid molecules which are to be analyzed are set upon a viscous medium, the gel, where an electric field induces the nucleic acids (which are negatively charged due to their sugar-phosphate backbone) to migrate toward the anode (which is positively charged because this is an electrolytic rather than galvanic cell). The separation of these fragments is accomplished by exploiting the mobilities with which different sized molecules are able to pass through the gel. Longer molecules migrate more slowly because they experience more resistance within the gel. Because the size of the molecule affects its mobility, smaller fragments end up nearer to the anode than longer ones in a given period. After some time, the voltage is removed and the fragmentation gradient is analyzed. For larger separations between similar sized fragments, either the voltage or run time ca ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
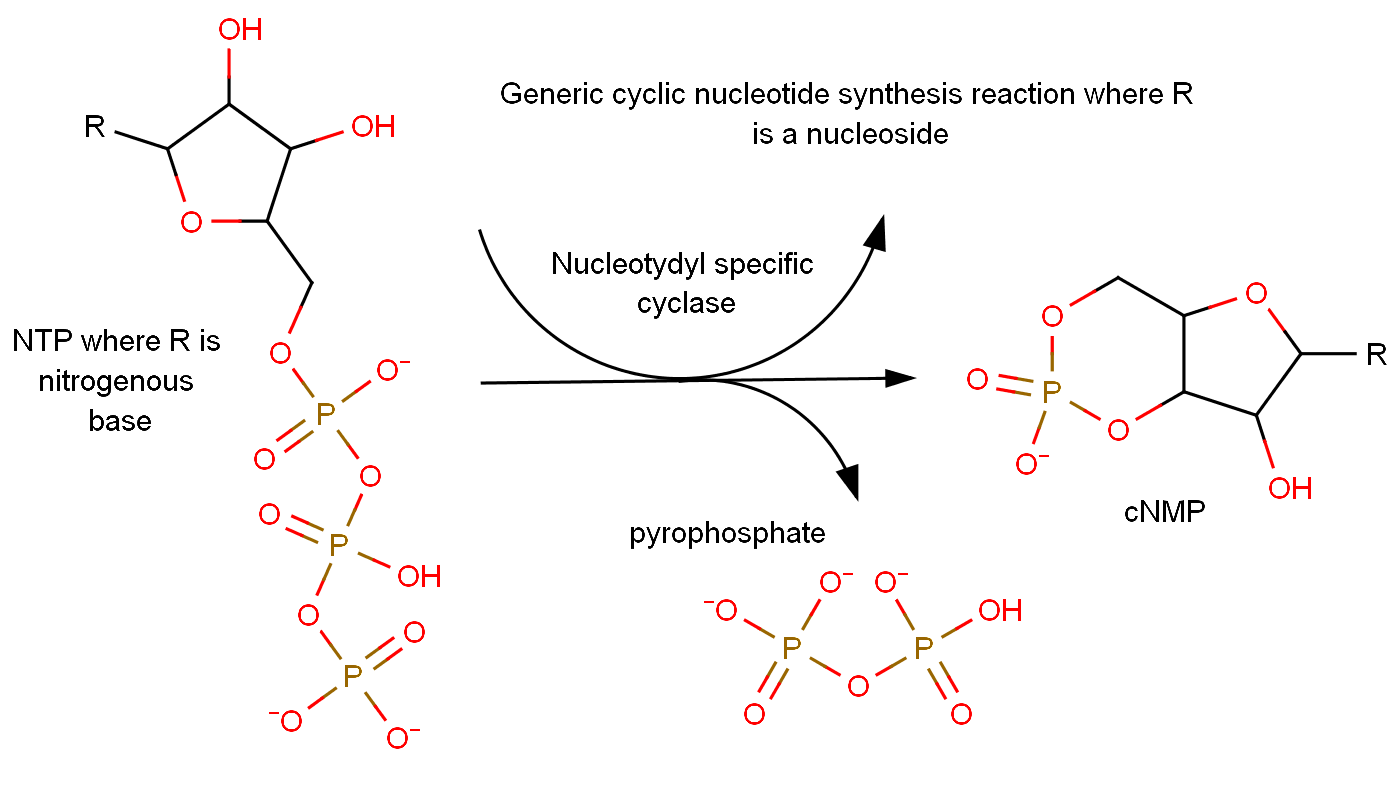
Cyclic Nucleotide
A cyclic nucleotide (cNMP) is a single-phosphate nucleotide with a cyclic bond arrangement between the sugar and phosphate groups. Like other nucleotides, cyclic nucleotides are composed of three functional groups: a sugar, a nitrogenous base, and a single phosphate group. As can be seen in the cyclic adenosine monophosphate (cAMP) and cyclic guanosine monophosphate (cGMP) images, the 'cyclic' portion consists of two bonds between the phosphate group and the 3' and 5' hydroxyl groups of the sugar, very often a ribose. Their biological significance includes a broad range of protein-ligand interactions. They have been identified as secondary messengers in both hormone and ion-channel signalling in eukaryotic cells, as well as allosteric effector compounds of DNA binding proteins in prokaryotic cells. cAMP and cGMP are currently the most well documented cyclic nucleotides, however there is evidence that cCMP (cytosine) is also involved in eukaryotic cellular messaging. The ro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cyclic Adenosine Monophosphate
Cyclic adenosine monophosphate (cAMP, cyclic AMP, or 3',5'-cyclic adenosine monophosphate) is a second messenger important in many biological processes. cAMP is a derivative of adenosine triphosphate (ATP) and used for intracellular signal transduction in many different organisms, conveying the cAMP-dependent pathway. History Earl Sutherland of Vanderbilt University won a Nobel Prize in Physiology or Medicine in 1971 "for his discoveries concerning the mechanisms of the action of hormones", especially epinephrine, via second messengers (such as cyclic adenosine monophosphate, cyclic AMP). Synthesis Cyclic adenosine monophosphate, AMP is synthesized from Adenosine triphosphate, ATP by adenylate cyclase located on the inner side of the plasma membrane and anchored at various locations in the interior of the cell. Adenylate cyclase is ''activated'' by a range of signaling molecules through the activation of adenylate cyclase stimulatory G (Gs alpha subunit, Gs)-protein-coupled recep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cyclic Guanosine Monophosphate
Cyclic guanosine monophosphate (cGMP) is a cyclic nucleotide derived from guanosine triphosphate (GTP). cGMP acts as a second messenger much like cyclic AMP. Its most likely mechanism of action is activation of intracellular protein kinases in response to the binding of membrane-impermeable peptide hormones to the external cell surface. Synthesis Guanylate cyclase (GC) catalyzes cGMP synthesis. This enzyme converts GTP to cGMP. Peptide hormones such as the atrial natriuretic factor activate membrane-bound GC, while soluble GC (sGC) is typically activated by nitric oxide to stimulate cGMP synthesis. sGC can be inhibited by ODQ (1H-,2,4xadiazolo ,3-auinoxalin-1-one). Functions cGMP is a common regulator of ion channel conductance, glycogenolysis, and cellular apoptosis. It also relaxes smooth muscle tissues. In blood vessels, relaxation of vascular smooth muscles leads to vasodilation and increased blood flow. At presynaptic terminals in the striatum, cGMP controls the e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
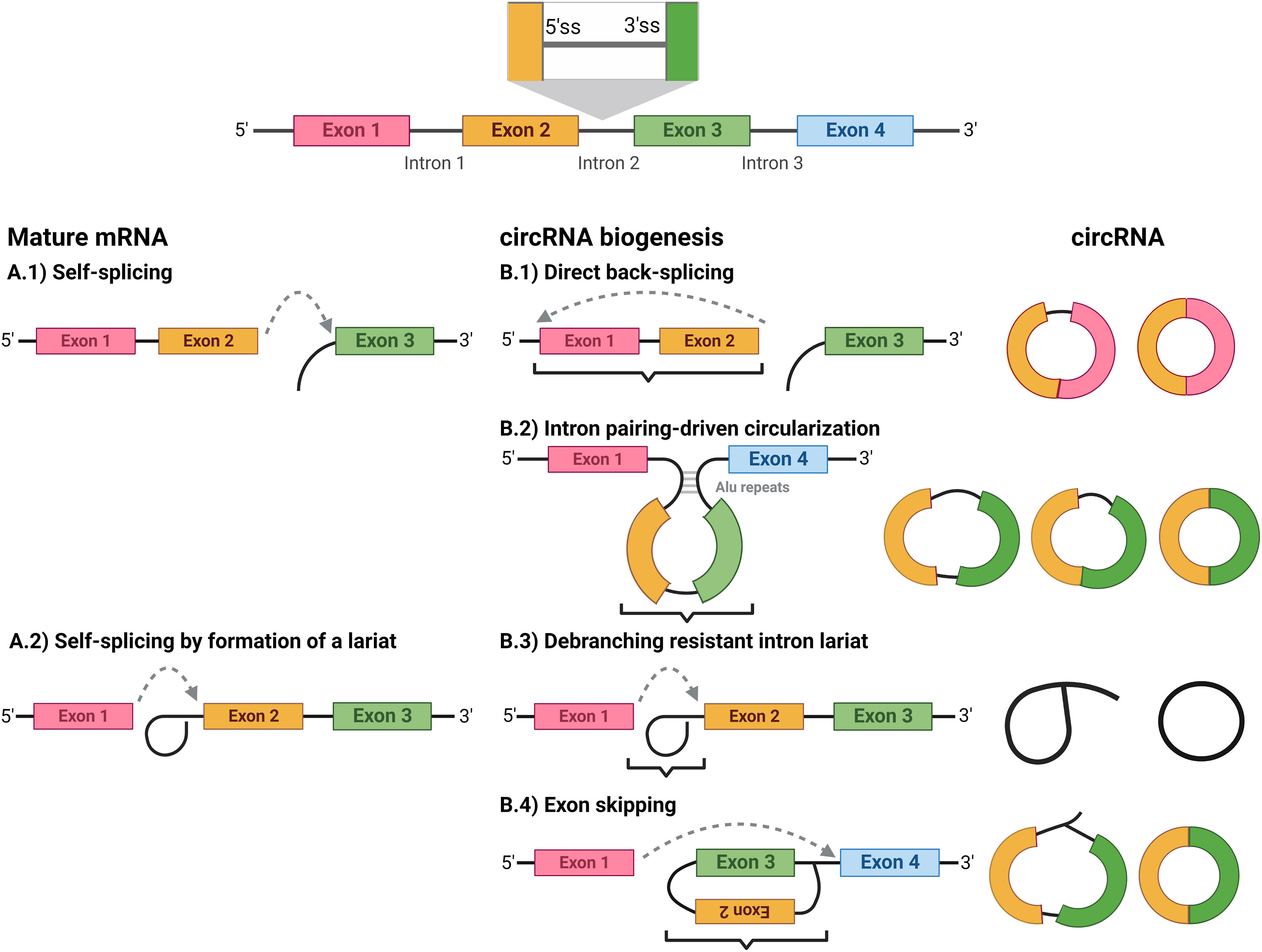
Circular RNA
Circular RNA (or circRNA) is a type of single-stranded RNA which, unlike linear RNA, forms a covalently closed continuous loop. In circular RNA, the 3' and 5' ends normally present in an RNA molecule have been joined together. This feature confers numerous properties to circular RNA, many of which have only recently been identified. Many types of circular RNA arise from otherwise protein-coding genes. Some circular RNA has been shown to code for proteins. Some types of circular RNA have also recently shown potential as gene regulators. The biological function of most circular RNA is unclear. Because circular RNA does not have 5' or 3' ends, it is resistant to exonuclease-mediated degradation and is presumably more stable than most linear RNA in cells. Circular RNA has been linked to some diseases such as cancer. RNA splicing In contrast to genes in bacteria, eukaryotic genes are split by non-coding sequences called introns. In eukaryotes, as a gene is transcribed from DNA int ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |