|
CREBBP
Cyclic adenosine monophosphate Response Element Binding protein Binding Protein (CREB-binding protein), also known as CREBBP or CBP or KAT3A, is a coactivator encoded by the ''CREBBP'' gene in humans, located on chromosome 16p13.3. CBP has intrinsic acetyltransferase functions; it is able to add acetyl groups to both transcription factors as well as histone lysines, the latter of which has been shown to alter chromatin structure making genes more accessible for transcription. This relatively unique acetyltransferase activity is also seen in another transcription enzyme, EP300 (p300). Together, they are known as the p300-CBP coactivator family and are known to associate with more than 16,000 genes in humans; however, while these proteins share many structural features, emerging evidence suggests that these two co-activators may promote transcription of genes with different biological functions. For example, CBP alone has been implicated in a wide variety of pathophysiologies includ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
KIX Domain
In biochemistry, the KIX domain (kinase-inducible domain (KID) interacting domain) or CREB binding domain is a protein domain of the eukaryotic transcriptional coactivators CBP and P300. It serves as a docking site for the formation of heterodimers between the coactivator and specific transcription factors. Structurally, the KIX domain is a globular domain consisting of three α-helices and two short 310-helices. The KIX domain was originally discovered in 1996 as the specific and minimal region in CBP that binds and interacts with phosphorylated CREB to activate transcription. It was thus first termed CREB-binding domain. However, when it was later discovered that it also binds many other proteins, the more general name KIX domain became favoured. The KIX domain contains two separate binding sites: the "c-Myb site", named after the oncoprotein c-Myb, and the "MLL site", named after the proto-oncogene MLL (Mixed Lineage Leukemia, KMT2A). The paralogous coactivators CBP (CREBBP ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
P300-CBP Coactivator Family
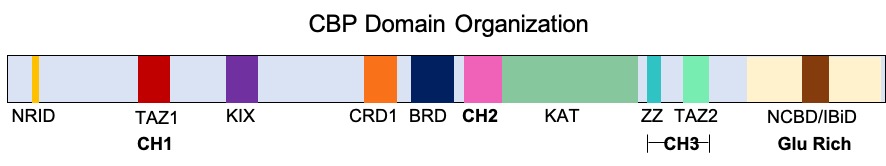
The p300-CBP coactivator family in humans is composed of two closely related transcriptional co-activating proteins (or coactivators): #p300 (also called EP300 or E1A binding protein p300) # CBP (also known as CREB-binding protein or CREBBP) Both p300 and CBP interact with numerous transcription factors and act to increase the expression of their target genes. Protein structure p300 and CBP have similar structures. Both contain five protein interaction domains: the nuclear receptor interaction domain (RID), the KIX domain (CREB and MYB interaction domain), the cysteine/histidine regions (TAZ1/CH1 and TAZ2/CH3) and the interferon response binding domain (IBiD). The last four domains, KIX, TAZ1, TAZ2 and IBiD of p300, each bind tightly to a sequence spanning both transactivation domains 9aaTADs of transcription factor p53. In addition p300 and CBP each contain a protein or histone acetyltransferase (PAT/HAT) domain and a bromodomain that binds acetylated lysines and a PHD ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
KMT2A
Histone-lysine ''N''-methyltransferase 2A also known as acute lymphoblastic leukemia 1 (ALL-1), myeloid/lymphoid or mixed-lineage leukemia 1 (MLL1), or zinc finger protein HRX (HRX) is an enzyme that in humans is encoded by the ''KMT2A'' gene. MLL1 is a histone methyltransferase deemed a positive global regulator of gene transcription. This protein belongs to the group of histone-modifying enzymes comprising transactivation domain 9aaTAD; ; and is involved in the epigenetic maintenance of transcriptional memory. Its role as an epigenetic regulator of neuronal function is an ongoing area of research. Function Transcriptional regulation KMT2A gene encodes a transcriptional coactivator that plays an essential role in regulating gene expression during early development and hematopoiesis. The encoded protein contains multiple conserved functional domains. One of these domains, the SET domain, is responsible for its histone H3 lysine 4 (H3K4) methyltransferase activity which medi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coactivator (genetics)
A coactivator is a type of transcriptional coregulator that binds to an activator (a transcription factor) to increase the rate of transcription of a gene or set of genes. The activator contains a DNA binding domain that binds either to a DNA promoter site or a specific DNA regulatory sequence called an enhancer. Binding of the activator-coactivator complex increases the speed of transcription by recruiting general transcription machinery to the promoter, therefore increasing gene expression. The use of activators and coactivators allows for highly specific expression of certain genes depending on cell type and developmental stage. Some coactivators also have histone acetyltransferase (HAT) activity. HATs form large multiprotein complexes that weaken the association of histones to DNA by acetylating the N-terminal histone tail. This provides more space for the transcription machinery to bind to the promoter, therefore increasing gene expression. Activators are found in all li ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CBP Domains
CBP may refer to: Business parks * Cebu Business Park, a central business district in Cebu City, Philippines * Changi Business Park, an eco-friendly industrial park in Singapore * Chiswick Business Park, a business park in Gunnersbury, West London Science and technology * Contention based protocol * CREB-binding protein a protein used in human transcriptional coactivation * Calcium-binding protein * Coded Block Pattern, a term used in video compression * Constrained Baseline Profile, the simplest of H.264/MPEG-4 AVC profiles Transport * Cangzhou West railway station, China Railway telegraph code CBP * Castle Bar Park railway station, National Rail station code CBP Other uses * Captive bolt pistol * Certified Benefits Professional, a certification for human-resource personnel * Chorleywood Bread Process * Citizens Bank Park, a baseball stadium used by the Philadelphia Phillies * Columbia Basin Project, a large irrigation network in central Washington * Crippled Black Phoenix, a Br ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Receptor Coactivator 1
The nuclear receptor coactivator 1 (''NCOA1'') is a transcriptional coregulatory protein that contains several nuclear receptor interacting domains and an intrinsic histone acetyltransferase activity. NCOA1 is recruited to DNA promotion sites by ligand-activated nuclear receptors. NCOA1, in turn, acylates histones, which makes downstream DNA more accessible to transcription. Hence, NCOA1 assists nuclear receptors in the upregulation of DNA expression. NCOA1 is also frequently called steroid receptor coactivator-1 (SRC-1). Interactions Nuclear receptor coactivator 1 possesses a basic helix-loop-helix (bHLH) domain and has been shown to interact with: * Androgen receptor, * C-Fos, * C-jun, * CIITA, * CREB-binding protein, * Cyclin D1, * DDX17, * DDX5 and * Estrogen receptor alpha, * Glucocorticoid receptor, * NFKB1, * PCAF, * PPARGC1A, * Peroxisome proliferator-activated receptor alpha, * SNW1, * STAT3, * STAT6, * TRIP4, and * Thyroid hormone receptor beta Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CREB
CREB-TF (CREB, cAMP response element-binding protein) is a cellular transcription factor. It binds to certain DNA sequences called cAMP response elements (CRE), thereby increasing or decreasing the transcription of the genes. CREB was first described in 1987 as a cAMP-responsive transcription factor regulating the somatostatin gene. Genes whose transcription is regulated by CREB include: '' c-fos'', BDNF, tyrosine hydroxylase, numerous neuropeptides (such as somatostatin, enkephalin, VGF, corticotropin-releasing hormone), and genes involved in the mammalian circadian clock (PER1, PER2). CREB is closely related in structure and function to CREM (cAMP response element modulator) and ATF-1 (activating transcription factor-1) proteins. CREB proteins are expressed in many animals, including humans. CREB has a well-documented role in neuronal plasticity and long-term memory formation in the brain and has been shown to be integral in the formation of spatial memory. CREB downreg ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MyoD
MyoD, also known as myoblast determination protein 1, is a protein in animals that plays a major role in regulating muscle differentiation. MyoD, which was discovered in the laboratory of Harold M. Weintraub, belongs to a family of proteins known as myogenic regulatory factors (MRFs). These bHLH (basic helix loop helix) transcription factors act sequentially in myogenic differentiation. Vertebrate MRF family members include MyoD1, Myf5, myogenin, and MRF4 (Myf6). In non-vertebrate animals, a single MyoD protein is typically found. MyoD is one of the earliest markers of myogenic commitment. MyoD is expressed at extremely low and essentially undetectable levels in quiescent satellite cells, but expression of MyoD is activated in response to exercise or muscle tissue damage. The effect of MyoD on satellite cells is dose-dependent; high MyoD expression represses cell renewal, promotes terminal differentiation and can induce apoptosis. Although MyoD marks myoblast commitment, musc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GATA1
GATA-binding factor 1 or GATA-1 (also termed Erythroid transcription factor) is the founding member of the GATA family of transcription factors. This protein is widely expressed throughout vertebrate species. In humans and mice, it is encoded by the ''GATA1'' and ''Gata1'' genes, respectively. These genes are located on the X chromosome in both species. GATA1 regulates the expression (i.e. formation of the genes' products) of an ensemble of genes that mediate the development of red blood cells and platelets. Its critical roles in red blood cell formation include promoting the maturation of precursor cells, e.g. erythroblasts, to red blood cells and stimulating these cells to erect their cytoskeleton and biosynthesize their oxygen-carrying components viz., hemoglobin and heme. GATA1 plays a similarly critical role in the maturation of blood platelets from megakaryoblasts, promegakaryocytes, and megakaryocytes; the latter cells then shed membrane-enclosed fragments of their cytop ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphorylation
In chemistry, phosphorylation is the attachment of a phosphate group to a molecule or an ion. This process and its inverse, dephosphorylation, are common in biology and could be driven by natural selection. Text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. Protein phosphorylation often activates (or deactivates) many enzymes. Glucose Phosphorylation of sugars is often the first stage in their catabolism. Phosphorylation allows cells to accumulate sugars because the phosphate group prevents the molecules from diffusing back across their transporter. Phosphorylation of glucose is a key reaction in sugar metabolism. The chemical equation for the conversion of D-glucose to D-glucose-6-phosphate in the first step of glycolysis is given by :D-glucose + ATP → D-glucose-6-phosphate + ADP : ΔG° = −16.7 kJ/mol (° indicates measurement at standard condition) Hepatic cells are freely permeable to glucose, and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
STAT3
Signal transducer and activator of transcription 3 (STAT3) is a transcription factor which in humans is encoded by the ''STAT3'' gene. It is a member of the STAT protein family. Function STAT3 is a member of the STAT protein family. In response to cytokines and growth factors, STAT3 is phosphorylated by receptor-associated Janus kinases (JAK), forms homo- or heterodimers, and translocates to the cell nucleus where it acts as a transcription activator. Specifically, STAT3 becomes activated after phosphorylation of tyrosine 705 in response to such ligands as interferons, epidermal growth factor (EGF), Interleukin (IL-)5 and IL-6. Additionally, activation of STAT3 may occur via phosphorylation of serine 727 by Mitogen-activated protein kinases (MAPK) and through c-src non-receptor tyrosine kinase. STAT3 mediates the expression of a variety of genes in response to cell stimuli, and thus plays a key role in many cellular processes such as cell growth and apoptosis. STAT3-deficien ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |