|
COX10
Protoheme IX farnesyltransferase, mitochondrial is an enzyme that in humans is encoded by the ''COX10'' gene. Cytochrome c oxidase (COX), the terminal component of the mitochondrial respiratory chain, catalyzes the electron transfer from reduced cytochrome c to oxygen. This component is a heteromeric complex consisting of 3 catalytic subunits encoded by mitochondrial genes and multiple structural subunits encoded by nuclear genes. The mitochondrially-encoded subunits function in electron transfer, and the nuclear-encoded subunits may function in the regulation and assembly of the complex. This nuclear gene, ''COX10'', encodes heme A: farnesyltransferase, which is not a structural subunit but required for the expression of functional COX and functions in the maturation of the heme A prosthetic group of COX. A gene mutation, which results in the substitution of a lysine for an asparagine (N204K), is identified to be responsible for cytochrome c oxidase deficiency. In addition ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytochrome C Oxidase
The enzyme cytochrome c oxidase or Complex IV, (was , now reclassified as a translocasEC 7.1.1.9 is a large transmembrane protein complex found in bacteria, archaea, and mitochondria of eukaryotes. It is the last enzyme in the respiratory electron transport chain of cells located in the membrane. It receives an electron from each of four cytochrome c molecules and transfers them to one oxygen molecule and four protons, producing two molecules of water. In addition to binding the four protons from the inner aqueous phase, it transports another four protons across the membrane, increasing the transmembrane difference of proton electrochemical potential, which the ATP synthase then uses to synthesize ATP. Structure The complex The complex is a large integral membrane protein composed of several metal prosthetic sites and 14 protein subunits in mammals. In mammals, eleven subunits are nuclear in origin, and three are synthesized in the mitochondria. The complex contains two ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication. The process of gene expression is used by all known life—eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and utilized by viruses—to generate the macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phenotype, '' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome. History The term ''exon'' derives from the expressed region and was coined by American biochemist Walter Gilbert in 1978: "The notion of the cistron… must be replaced by that of a transcription unit containing regions which will be lost from the mature messengerwhich I suggest we call introns (for intragenic regions)alternating with regions which will be expressedexons." This definition was originally made for protein-coding transcripts that are spliced before being translated. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Open Reading Frame
In molecular biology, open reading frames (ORFs) are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames will be "open" (the "reading", however, refers to the RNA produced by transcription of the DNA and its subsequent interaction with the ribosome in translation). Such an ORF may contain a start codon (usually AUG in terms of RNA) and by definition cannot extend beyond a stop codon (usually UAA, UAG or UGA in RNA). That start codon (not necessarily the first) indicates where translation may start. The transcription termination site is located after the ORF, beyond the translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In eukaryotic genes with multiple exons, introns are removed and exons are then joined together after transcription to yield the final ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Three Prime Untranslated Region
In molecular genetics, the three prime untranslated region (3′-UTR) is the section of messenger RNA (mRNA) that immediately follows the translation termination codon. The 3′-UTR often contains regulatory regions that post-transcriptionally influence gene expression. During gene expression, an mRNA molecule is transcribed from the DNA sequence and is later translated into a protein. Several regions of the mRNA molecule are not translated into a protein including the 5' cap, 5' untranslated region, 3′ untranslated region and poly(A) tail. Regulatory regions within the 3′-untranslated region can influence polyadenylation, translation efficiency, localization, and stability of the mRNA. The 3′-UTR contains both binding sites for regulatory proteins as well as microRNAs (miRNAs). By binding to specific sites within the 3′-UTR, miRNAs can decrease gene expression of various mRNAs by either inhibiting translation or directly causing degradation of the transcript. The 3� ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acids
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha amino acids appear in the genetic code. Amino acids can be classified according to the locations of the core structural functional groups, as Alpha and beta carbon, alpha- , beta- , gamma- or delta- amino acids; other categories relate to Chemical polarity, polarity, ionization, and side chain group type (aliphatic, Open-chain compound, acyclic, aromatic, containing hydroxyl or sulfur, etc.). In the form of proteins, amino acid '' residues'' form the second-largest component (water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling lif ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromosome 17
Chromosome 17 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 17 spans more than 83 million base pairs (the building material of DNA) and represents between 2.5 and 3% of the total DNA in cells. Chromosome 17 contains the Homeobox B gene cluster. Genes Number of genes The following are some of the gene count estimates of human chromosome 17. Because researchers use different approaches to genome annotation their predictions of the number of genes on each chromosome varies (for technical details, see gene prediction). Among various projects, the collaborative consensus coding sequence project ( CCDS) takes an extremely conservative strategy. So CCDS's gene number prediction represents a lower bound on the total number of human protein-coding genes. Gene list The following is a partial list of genes on human chromosome 17. For complete list, see the link in the infobox on the right. The following are some o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
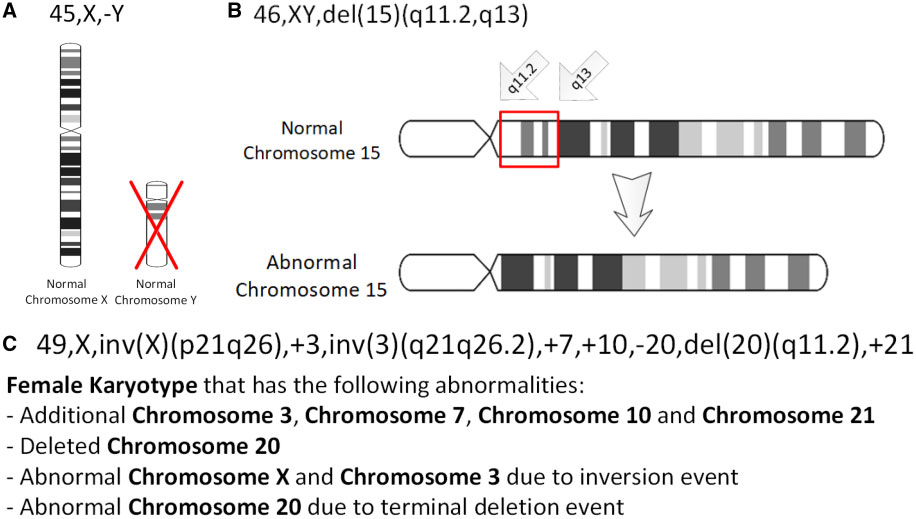
Deletion (genetics)
In genetics, a deletion (also called gene deletion, deficiency, or deletion mutation) (sign: Δ) is a mutation (a genetic aberration) in which a part of a chromosome or a sequence of DNA is left out during DNA replication. Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome. Some chromosomes have fragile spots where breaks occur which result in the deletion of a part of chromosome. The breaks can be induced by heat, viruses, radiations, chemicals. When a chromosome breaks, a part of it is deleted or lost, the missing piece of chromosome is referred to as deletion or a deficiency. For synapsis to occur between a chromosome with a large intercalary deficiency and a normal complete homolog, the unpaired region of the normal homolog must loop out of the linear structure into a deletion or compensation loop. The smallest single base deletion mutations occur by a single base flipping in the template DNA, followed by template DNA strand sli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hereditary Neuropathy With Liability To Pressure Palsy
Hereditary neuropathy with liability to pressure palsy (HNPP) is a peripheral neuropathy, a condition that affects the nerves.update 2014 Pressure on the nerves can cause tingling sensations, numbness, pain, weakness, muscle atrophy and even paralysis of the affected area. In normal individuals, these symptoms disappear quickly, but in sufferers of HNPP even a short period of pressure can cause the symptoms to occur. Palsies can last from minutes or days to weeks or even months. HNPP is caused by a mutation in the gene ''PMP22'', which makes peripheral myelin protein 22. This protein has a role in the maintenance of the myelin sheath that insulates nerves, resulting in insufficient conductivity in the nerves. HNPP is part of the group of hereditary motor and sensory neuropathy (HMSN) disorders and is linked to Charcot–Marie–Tooth disease (CMT). Signs and symptoms Symptoms and symptom onset vary; some individuals are diagnosed in childhood, others in adulthood, some report mino ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Charcot–Marie–Tooth Disease Classifications
Classifications of Charcot–Marie–Tooth disease refers to the types and subtypes of Charcot–Marie–Tooth disease (CMT), a genetically and clinically heterogeneous group of inherited disorders of the peripheral nervous system characterized by progressive loss of muscle tissue and touch sensation across various parts of the body. CMT is a result of genetic mutations in a number of gene In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a ba ...s. __TOC__ Clinical categories Genetic subtypes It has to be kept in mind that sometimes a particular patient diagnosed with CMT can exhibit a combination of any of the above gene mutations; thus, in these cases precise classification can be arbitrary. References {{DEFAULTSORT:Charcot-Marie-Tooth disease classifications Peripheral ne ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Charcot–Marie–Tooth Disease
Charcot–Marie–Tooth disease (CMT) is a hereditary motor and sensory neuropathy of the peripheral nervous system characterized by progressive loss of muscle tissue and touch sensation across various parts of the body. This disease is the most commonly inherited neurological disorder, affecting about one in 2,500 people. It is named after those who classically described it: the Frenchman Jean-Martin Charcot (1825–1893), his pupil Pierre Marie (1853–1940), and the Briton Howard Henry Tooth (1856–1925). There is no known cure. Care focuses on maintaining function. CMT was previously classified as a subtype of muscular dystrophy. Signs and symptoms Symptoms of CMT usually begin in early childhood or early adulthood but can begin later. Some people do not experience symptoms until their early 30s or 40s. Usually, the initial symptom is foot drop early in the course of the disease. This can also cause hammertoe, where the toes are always curled. Wasting of muscle tissue of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |