|
Protein Engineering
Protein engineering is the process of developing useful or valuable proteins. It is a young discipline, with much research taking place into the understanding of protein folding and recognition for protein design principles. It has been used to improve the function of many enzymes for industrial catalysis. It is also a product and services market, with an estimated value of $168 billion by 2017. There are two general strategies for protein engineering: rational protein design and directed evolution. These methods are not mutually exclusive; researchers will often apply both. In the future, more detailed knowledge of protein structure and function, and advances in high-throughput screening, may greatly expand the abilities of protein engineering. Eventually, even unnatural amino acids may be included, via newer methods, such as expanded genetic code, that allow encoding novel amino acids in genetic code. Approaches Rational design In rational protein design, a scientist uses ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UPGMA
UPGMA (unweighted pair group method with arithmetic mean) is a simple agglomerative (bottom-up) hierarchical clustering method. The method is generally attributed to Sokal and Michener. The UPGMA method is similar to its ''weighted'' variant, the WPGMA method. Note that the unweighted term indicates that all distances contribute equally to each average that is computed and does not refer to the math by which it is achieved. Thus the simple averaging in WPGMA produces a weighted result and the proportional averaging in UPGMA produces an unweighted result ('' see the working example''). Algorithm The UPGMA algorithm constructs a rooted tree (dendrogram) that reflects the structure present in a pairwise similarity matrix (or a dissimilarity matrix). At each step, the nearest two clusters are combined into a higher-level cluster. The distance between any two clusters \mathcal and \mathcal, each of size (''i.e.'', cardinality) and , is taken to be the average of all distances d(x,y) ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Homology
Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal (or lateral) gene transfer event (xenologs). Homology among DNA, RNA, or proteins is typically inferred from their nucleotide or amino acid sequence similarity. Significant similarity is strong evidence that two sequences are related by evolutionary changes from a common ancestral sequence. Alignments of multiple sequences are used to indicate which regions of each sequence are homologous. Identity, similarity, and conservation The term "percent homology" is often used to mean "sequence similarity”, that is the percentage of identical residues (''percent identity''), or the percentage of residues conserved with similar physicochemical properties (' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Saturation Mutagenesis
Sequence saturation mutagenesis (SeSaM) is a chemo-enzymatic random mutagenesis method applied for the directed evolution of proteins and enzymes. It is one of the most common saturation mutagenesis techniques. In four PCR-based reaction steps, phosphorothioate nucleotides are inserted in the gene sequence, cleaved and the resulting fragments elongated by universal or degenerate nucleotides. These nucleotides are then replaced by standard nucleotides, allowing for a broad distribution of nucleic acid mutations spread over the gene sequence with a preference to transversions and with a unique focus on consecutive point mutations, both difficult to generate by other mutagenesis techniques. The technique was developed by Professor Ulrich Schwaneberg at Jacobs University Bremen and RWTH Aachen University. Technology, development and advantages SeSaM has been developed in order to overcome several of the major limitations encountered when working with standard mutagenesis methods bas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies (complete or partial) of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it (or a part of it) to a large enough amount to study in detail. PCR was invented in 1983 by the American biochemist Kary Mullis at Cetus Corporation; Mullis and biochemist Michael Smith (chemist), Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of Ancient DNA, ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Recombinant DNA
Recombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods of genetic recombination (such as molecular cloning) that bring together genetic material from multiple sources, creating sequences that would not otherwise be found in the genome. Recombinant DNA is the general name for a piece of DNA that has been created by combining at least two fragments from two different sources. Recombinant DNA is possible because DNA molecules from all organisms share the same chemical structure, and differ only in the nucleotide sequence within that identical overall structure. Recombinant DNA molecules are sometimes called chimeric DNA, because they can be made of material from two different species, like the mythical chimera. R-DNA technology uses palindromic sequences and leads to the production of sticky and blunt ends. The DNA sequences used in the construction of recombinant DNA molecules can originate from any species. For example, plant DNA may be joined to bacter ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sexual Reproduction
Sexual reproduction is a type of reproduction that involves a complex life cycle in which a gamete ( haploid reproductive cells, such as a sperm or egg cell) with a single set of chromosomes combines with another gamete to produce a zygote that develops into an organism composed of cells with two sets of chromosomes ( diploid). This is typical in animals, though the number of chromosome sets and how that number changes in sexual reproduction varies, especially among plants, fungi, and other eukaryotes. Sexual reproduction is the most common life cycle in multicellular eukaryotes, such as animals, fungi and plants. Sexual reproduction also occurs in some unicellular eukaryotes. Sexual reproduction does not occur in prokaryotes, unicellular organisms without cell nuclei, such bacteria and archaea. However, some process in bacteria may be considered analogous to sexual reproduction in that they incorporate new genetic information, including bacterial conjugation, transformatio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homologous Recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in cellular organisms but may be also RNA in viruses). Homologous recombination is widely used by cells to accurately DNA repair harmful breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses. Horizontal ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
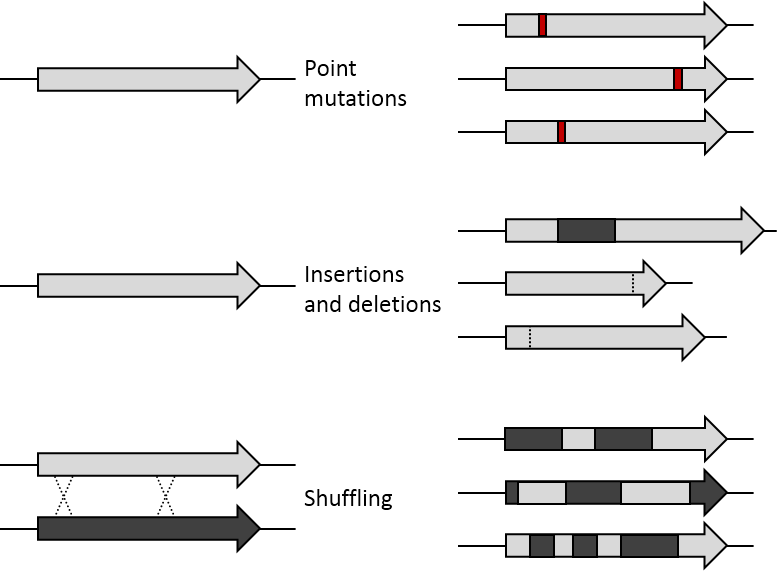
DNA Shuffling
DNA shuffling, also known as molecular breeding, is an in vitro random recombination method to generate mutant genes for directed evolution and to enable a rapid increase in DNA library size. Three procedures for accomplishing DNA shuffling are molecular breeding which relies on homologous recombination or the similarity of the DNA sequences, restriction enzymes which rely on common restriction sites, and nonhomologous random recombination which requires the use of hairpins. In all of these techniques, the parent genes are fragmented and then recombined. DNA shuffling utilizes random recombination as opposed to site-directed mutagenesis in order to generate proteins with unique attributes or combinations of desirable characteristics encoded in the parent genes such as thermostability and high activity. The potential for DNA shuffling to produce novel proteins is exemplified by the figure shown on the right which demonstrates the difference between point mutations, insertions and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Evolution
Evolution is change in the heritable characteristics of biological populations over successive generations. These characteristics are the expressions of genes, which are passed on from parent to offspring during reproduction. Variation tends to exist within any given population as a result of genetic mutation and recombination. Evolution occurs when evolutionary processes such as natural selection (including sexual selection) and genetic drift act on this variation, resulting in certain characteristics becoming more common or more rare within a population. The evolutionary pressures that determine whether a characteristic is common or rare within a population constantly change, resulting in a change in heritable characteristics arising over successive generations. It is this process of evolution that has given rise to biodiversity at every level of biological organisation, including the levels of species, individual organisms, and molecules. The theory of evolution by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Saturation Mutagenesis
Sequence saturation mutagenesis (SeSaM) is a chemo-enzymatic random mutagenesis method applied for the directed evolution of proteins and enzymes. It is one of the most common saturation mutagenesis techniques. In four PCR-based reaction steps, phosphorothioate nucleotides are inserted in the gene sequence, cleaved and the resulting fragments elongated by universal or degenerate nucleotides. These nucleotides are then replaced by standard nucleotides, allowing for a broad distribution of nucleic acid mutations spread over the gene sequence with a preference to transversions and with a unique focus on consecutive point mutations, both difficult to generate by other mutagenesis techniques. The technique was developed by Professor Ulrich Schwaneberg at Jacobs University Bremen and RWTH Aachen University. Technology, development and advantages SeSaM has been developed in order to overcome several of the major limitations encountered when working with standard mutagenesis methods bas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |