|
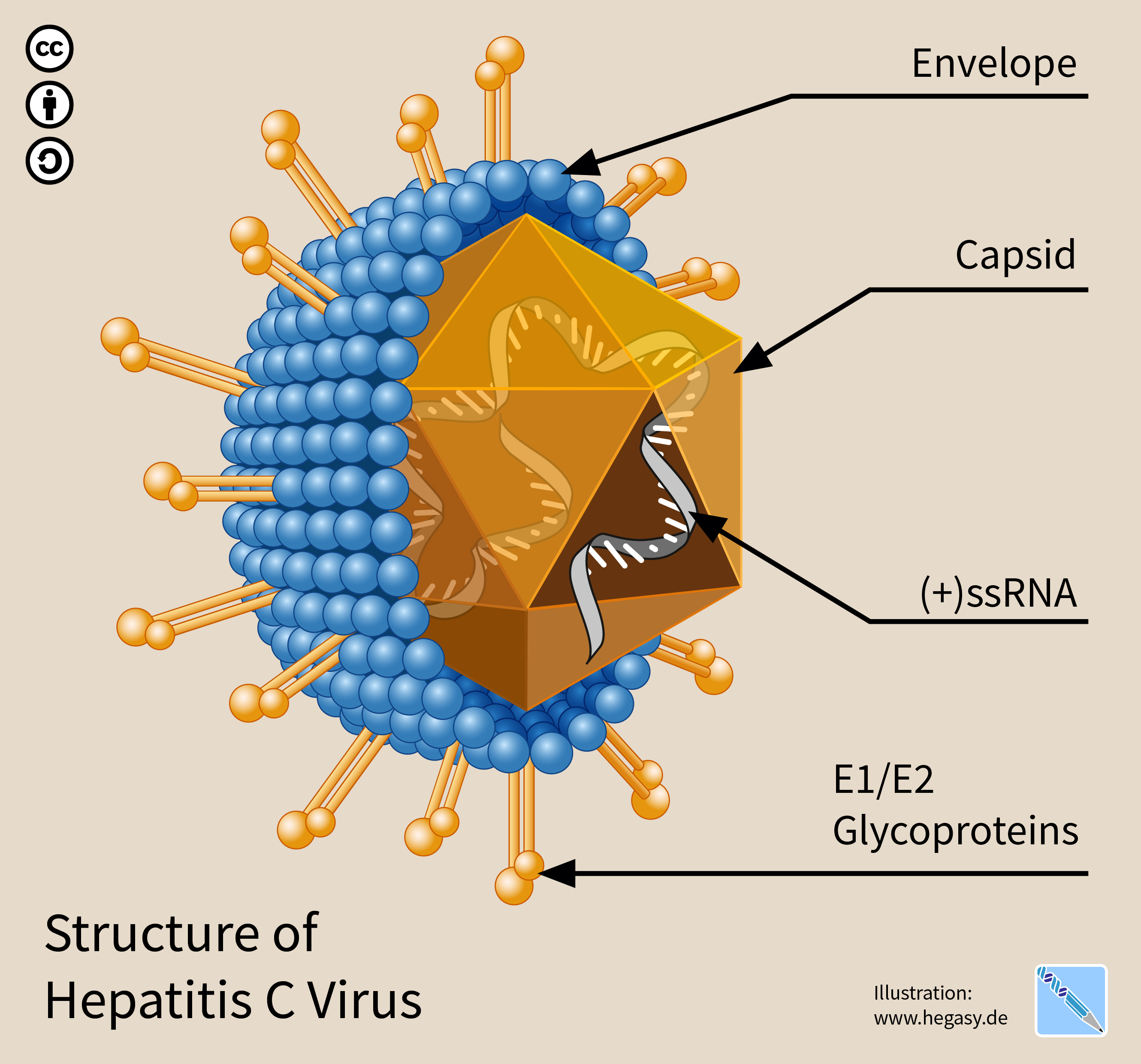
Hepatitis C Virus
The hepatitis C virus (HCV) is a small (55–65 nm in size), enveloped, positive-sense single-stranded RNA virus of the family ''Flaviviridae''. The hepatitis C virus is the cause of hepatitis C and some cancers such as liver cancer ( hepatocellular carcinoma, abbreviated HCC) and lymphomas in humans. Taxonomy The hepatitis C virus belongs to the genus '' Hepacivirus'', a member of the family ''Flaviviridae''. Before 2011, it was considered to be the only member of this genus. However a member of this genus has been discovered in dogs: canine hepacivirus. There is also at least one virus in this genus that infects horses. Several additional viruses in the genus have been described in bats and rodents. Structure The hepatitis C virus particle consists of a lipid membrane envelope that is 55 to 65 nm in diameter. Two viral envelope glycoproteins, E1 and E2, are embedded in the lipid envelope. They take part in viral attachment and entry into the cell. Within the e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electron Micrograph
A micrograph is an image, captured photographically or digitally, taken through a microscope or similar device to show a magnify, magnified image of an object. This is opposed to a macrograph or photomacrograph, an image which is also taken on a microscope but is only slightly magnified, usually less than 10 times. Micrography is the practice or art of using microscopes to make photographs. A photographic micrograph is a photomicrograph, and one taken with an electron microscope is an electron micrograph. A micrograph contains extensive details of microstructure. A wealth of information can be obtained from a simple micrograph like behavior of the material under different conditions, the phases found in the system, failure analysis, grain size estimation, elemental analysis and so on. Micrographs are widely used in all fields of microscopy. Types Photomicrograph A light micrograph or photomicrograph is a micrograph prepared using an optical microscope, a process referred to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
E1 (HCV)
E1 is one of two subunits of the envelope glycoprotein found in the hepatitis C virus. The other subunit is E2. This protein is a type 1 transmembrane protein with a highly glycosylated N-terminal ectodomain and a C-terminal hydrophobic anchor. After being synthesized the E1 glycoproteins associates with the E2 glycoprotein as a noncovalent heterodimer. Structure The E1 glycoprotein residues 192-383 in the genotype 1a H77 strain. After translation the E1 C-terminal transmembrane domains (TMDs) forms a hairpin of antiparallel a-helices. E1 is then cleaved by signal peptide peptidase at the endoplasmic reticulum and E1 is then made into a single long straight a-helix. What is known of the structure is from a crystal structure made in 2014. This crystal structure shows that it has two a-helixes and 3 B-sheets for both monomers; two disulfide bridges stabilize these two monomers. This means that E1 is more compact then its E2 counterpart. It has been shown that E1 can fold with a sma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
European Bioinformatics Institute
The European Bioinformatics Institute (EMBL-EBI) is an intergovernmental organization (IGO) which, as part of the European Molecular Biology Laboratory (EMBL) family, focuses on research and services in bioinformatics. It is located on the Wellcome Genome Campus in Hinxton near Cambridge, and employs over 600 full-time equivalent (FTE) staff. Further, the EMBL-EBI hosts training programs that teach scientists the fundamentals of the work with biological data and promote the plethora of bioinformatic tools available for their research, both EMBL-EBI-based and not so. Bioinformatic services One of the roles of the EMBL-EBI is to index and maintain biological data in a set of databases, including Ensembl (housing whole genome sequence data), UniProt (protein sequence and annotation database) and Protein Data Bank (protein and nucleic acid tertiary structure database). A variety of online services and tools is provided, such as Basic Local Alignment Search Tool (BLAST) or Clust ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleotide
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all Life, life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common Nutrient, nutrients by the liver. Nucleotides are composed of three subunit molecules: a nucleobase, a pentose, five-carbon sugar (ribose or deoxyribose), and a phosphate group consisting of one to three phosphates. The four nucleobases in DNA are guanine, adenine, cytosine, and thymine; in RNA, uracil is used in place of thymine. Nucleotides also play a central role in metabolism at a fundamental, cellular level. They provide chemical energy—in the form of the nucleoside triphosphates, adenosine triphosphate (ATP), guanosine triphosphate (GTP), cytidine triphosphate (CTP), and uridine triph ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Open Reading Frame
In molecular biology, reading frames are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames will be "open" (the "reading", however, refers to the RNA produced by transcription of the DNA and its subsequent interaction with the ribosome in translation). Such an open reading frame (ORF) may contain a start codon (usually AUG in terms of RNA) and by definition cannot extend beyond a stop codon (usually UAA, UAG or UGA in RNA). That start codon (not necessarily the first) indicates where translation may start. The transcription termination site is located after the ORF, beyond the translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In eukaryotic genes with multiple exons, introns are removed and exons are then joined together after transcription to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences (see non-coding DNA), and often a substantial fraction of junk DNA with no evident function. Almost all eukaryotes have mitochondrial DNA, mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplast DNA, chloroplasts with a chloroplast genome. The study of the genome is called genomics. The genomes of many organisms have been Whole-genome sequencing, sequenced and various regions have been annotated. The first genome to be sequenced was that of the virus φX174 in 1977; the first genome sequence of a prokaryote (''Haemophilus influenzae'') was published in 1995; the yeast (''Saccharomyces cerevisiae'') genome was the first eukaryotic genome to be sequenced in 1996. The Human Genome Project ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-stranded
When referring to DNA transcription, the coding strand (or informational strand) is the DNA strand whose base sequence is identical to the base sequence of the RNA transcript produced (although with thymine replaced by uracil). It is this strand which contains codons, while the non-coding strand contains anticodons. During transcription, RNA Pol II binds to the non-coding template strand, reads the anti-codons, and transcribes their sequence to synthesize an RNA transcript with complementary bases. By convention, the coding strand is the strand used when displaying a DNA sequence. It is presented in the 5' to 3' direction. Wherever a gene exists on a DNA molecule, one strand is the coding strand (or sense strand), and the other is the noncoding strand (also called the antisense strand, anticoding strand, template strand or transcribed strand). Strands in transcription bubble During transcription, RNA polymerase unwinds a short section of the DNA double helix near the s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Positive Sense
In molecular biology and genetics, the sense of a nucleic acid molecule, particularly of a strand of DNA or RNA, refers to the nature of the roles of the strand and its complement in specifying a sequence of amino acids. Depending on the context, sense may have slightly different meanings. For example, the negative-sense strand of DNA is equivalent to the template strand, whereas the positive-sense strand is the non-template strand whose nucleotide sequence is equivalent to the sequence of the mRNA transcript. DNA sense Because of the complementary nature of base-pairing between nucleic acid polymers, a double-stranded DNA molecule will be composed of two strands with sequences that are reverse complements of each other. To help molecular biologists specifically identify each strand individually, the two strands are usually differentiated as the "sense" strand and the "antisense" strand. An individual strand of DNA is referred to as positive-sense (also positive (+) or simply sen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HCV IRES
The Hepatitis C virus internal ribosome entry site, or HCV IRES, is an RNA structure within the 5'UTR of the HCV genome that mediates cap-independent translation initiation. Protein translation of most eukaryotic mRNAs occurs by a cap-dependent mechanism and requires association of Met-tRNAiMet, several eukaryotic initiation factors, and GTP with the 40S ribosomal subunit, recruitment to the 5' cap, and scanning along the 5' UTR to reach to start codon. In contrast, translation of hepatitis C virus (HCV) mRNA is initiated by a different mechanism from the usual 5' cap-binding model. This alternate mechanism relies on the direct binding of the 40S ribosomal subunit by the internal ribosome entry site (IRES) in the 5' UTR of HCV RNA. The HCV IRES adopts a complex structure, and may differ significantly from IRES elements identified in picornaviruses. A small number of eukaryotic mRNAs has been shown to be translated by internal ribosome entry. IRES structure Nucleotides 1� ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CD81
CD81 molecule, also known as CD81 (Cluster of Differentiation 81), is a protein which in humans is encoded by the ''CD81'' gene. It is also known as 26 kDa cell surface protein, TAPA-1 (Target of the Antiproliferative Antibody 1), and Tetraspanin-28 (Tspan-28). Gene The gene is located on the plus strand of the short arm of chromosome 11 (11p15.5). It is 20,103 bases in length and encodes a protein of 236 amino acids (predicted molecular weight 25.809 kDa). The protein does not appear to be post translationally modified and has four transmembrane domains. Both the N-terminus and C-terminus lie on the intracellular side of the membrane. The gene is expressed in hemopoietic, endothelial, and epithelial cells. It is absent from erythrocytes, platelets, and neutrophils. Function The protein encoded by this gene is a member of the transmembrane 4 superfamily, also known as the tetraspanin family. Most of these members are cell-surface proteins that are characterized by th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hypervariable Region
A hypervariable region (HVR) is a location within a sequence where polymorphisms frequently occur. It is used in two contexts: * In the case of nucleic acids, an HVR is where base pairs frequently change. This can be due to a change in the number of repeats (which is seen in eukaryotic nuclear DNA) or simply low selective pressure allowing a great number of substitutions and indels (as in the case of mitochondrial DNA D-loop and 16S rRNA). * In the case of antibodies, an HVR is where most of the differences among antibodies occur. This region is also called the complementarity-determining region. Because there already is a separate article for the antibody region, this article will focus on the nucleic acid case. Mitochondrial There are two mitochondrial hypervariable regions used in human mitochondrial genealogical DNA testing. HVR1 is considered a "low resolution" region and HVR2 is considered a "high resolution" region. Getting HVR1 and HVR2 DNA tests can help determine one ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Disulfide Bonds
In chemistry, a disulfide (or disulphide in British English) is a compound containing a functional group or the anion. The linkage is also called an SS-bond or sometimes a disulfide bridge and usually derived from two thiol groups. In inorganic chemistry, the anion appears in a few rare minerals, but the functional group has tremendous importance in biochemistry. Disulfide bridges formed between thiol groups in two cysteine residues are an important component of the tertiary and quaternary structure of proteins. Compounds of the form are usually called '' persulfides'' instead. Organic disulfides Structure Disulfides have a C–S–S–C dihedral angle approaching 90°. The S–S bond length is 2.03 Å in diphenyl disulfide, similar to that in elemental sulfur. Disulfides are usually symmetric but they can also be unsymmetric. Symmetrical disulfides are compounds of the formula . Most disulfides encountered in organosulfur chemistry are symmetrical disulfides. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |