|
EC 1.13
This list contains a list of EC numbers for the first group, EC 1, oxidoreductases, placed in numerical order as determined by the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology. All official information is tabulated at the website of the committee. The database is developed and maintained by Andrew McDonald. EC 1.1 Acting on the CH-OH group of donors EC 1.1.1 With Nicotinamide adenine dinucleotide or NADP as acceptor *: alcohol dehydrogenase *: alcohol dehydrogenase (NADP+) *: homoserine dehydrogenase *: (''R,R'')-butanediol dehydrogenase * EC 1.1.1.5: acetoin dehydrogenase. Now EC 1.1.1.303, diacetyl reductase ''R'')-acetoin formingand EC 1.1.1.304, diacetyl reductase ''S'')-acetoin forming*: glycerol dehydrogenase *: propanediol-phosphate dehydrogenase *: glycerol-3-phosphate dehydrogenase (NAD+) *: D-xylulose reductase *: L-xylulose reductase *: D-arabinitol 4-dehydrogenase *: L-arabinitol 4-dehydrogenase *: L-arabinitol 2-d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme Commission Number
The Enzyme Commission number (EC number) is a numerical classification scheme for enzymes, based on the chemical reactions they catalyze. As a system of enzyme nomenclature, every EC number is associated with a recommended name for the corresponding enzyme-catalyzed reaction. EC numbers do not specify enzymes but enzyme-catalyzed reactions. If different enzymes (for instance from different organisms) catalyze the same reaction, then they receive the same EC number. Furthermore, through convergent evolution, completely different protein folds can catalyze an identical reaction (these are sometimes called non-homologous isofunctional enzymes) and therefore would be assigned the same EC number. By contrast, UniProt identifiers uniquely specify a protein by its amino acid sequence. Format of number Every enzyme code consists of the letters "EC" followed by four numbers separated by periods. Those numbers represent a progressively finer classification of the enzyme. Preliminary EC ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Galactitol 2-dehydrogenase
In enzymology, a galactitol 2-dehydrogenase () is an enzyme that catalyzes the chemical reaction :galactitol + NAD+ \rightleftharpoons D-tagatose + NADH + H+ Thus, the two substrates of this enzyme are galactitol and NAD+, whereas its 3 products are D-tagatose, NADH, and H+. This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-OH group of donor with NAD+ or NADP+ as acceptor. The systematic name of this enzyme class is galactitol:NAD+ 2-oxidoreductase. This enzyme is also called dulcitol dehydrogenase. This enzyme participates in galactose metabolism Galactose (, '' galacto-'' + ''-ose'', "milk sugar"), sometimes abbreviated Gal, is a monosaccharide sugar that is about as sweet as glucose, and about 65% as sweet as sucrose. It is an aldohexose and a C-4 epimer of glucose. A galactose mole .... References * EC 1.1.1 NADH-dependent enzymes Enzymes of known structure {{1.1.1-enzyme-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glyoxylate Reductase
Glyoxylate reductase (), first isolated from spinach leaves, is an enzyme that catalyzes the reduction of glyoxylate to glycolate, using the cofactor NADH or NADPH. The systematic name of this enzyme class is glycolate:NAD+ oxidoreductase. Other names in common use include NADH-glyoxylate reductase, glyoxylic acid reductase, and NADH-dependent glyoxylate reductase. Structure The crystal structure of the glyoxylate reductase enzyme from the hyperthermophilic archeon Pyrococcus horiskoshii OT3 has been reported. The enzyme exists in the dimeric form. Each monomer has two domains: a substrate- binding domain where glyoxylate binds, and a nucleotide-binding domain where the NAD(P)H cofactor binds. Mechanism The enzyme catalyzes the transfer of a hydride from NAD(P)H to glyoxylate, causing a reduction of the substrate to glycolate and an oxidation of the cofactor to NAD(P)+. Figure 2 shows the mechanism for this reaction. It is thought that the two residues Glu270 and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Shikimate Dehydrogenase
In enzymology, a shikimate dehydrogenase () is an enzyme that catalyzes the chemical reaction :shikimate + NADP+ \rightleftharpoons 3-dehydroshikimate + NADPH + H+ Thus, the two substrates of this enzyme are shikimate and NADP+, whereas its 3 products are 3-dehydroshikimate, NADPH, and H+. This enzyme participates in phenylalanine, tyrosine and tryptophan biosynthesis. Function Shikimate dehydrogenase is an enzyme that catalyzes one step of the shikimate pathway. This pathway is found in bacteria, plants, fungi, algae, and parasites and is responsible for the biosynthesis of aromatic amino acids (phenylalanine, tyrosine, and tryptophan) from the metabolism of carbohydrates. In contrast, animals and humans lack this pathway hence products of this biosynthetic route are essential amino acids that must be obtained through an animal's diet. There are seven enzymes that play a role in this pathway. Shikimate dehydrogenase (also known as 3-dehydroshikimate dehydrogenase) is t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Quinate Dehydrogenase
In enzymology, a quinate dehydrogenase () is an enzyme that catalyzes the chemical reaction :L-quinate + NAD+ \rightleftharpoons 3-dehydroquinate + NADH + H+ Thus, the two substrates of this enzyme are L-quinate and NAD+, whereas its 3 products Product may refer to: Business * Product (business), an item that serves as a solution to a specific consumer problem. * Product (project management), a deliverable or set of deliverables that contribute to a business solution Mathematics * Produ ... are 3-dehydroquinate, nicotinamide adenine dinucleotide, NADH, and hydrogen ion, H+. This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-OH group of donor with NAD+ or NADP+ as acceptor. The List of enzymes, systematic name of this enzyme class is L-quinate:NAD+ 3-oxidoreductase. Other names in common use include quinic dehydrogenase, quinate:NAD oxidoreductase, quinate 5-dehydrogenase, and quinate:NAD+ 5-oxidoreductase. This enzyme participates in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histidinol Dehydrogenase
In enzymology, histidinol dehydrogenase (HIS4) (HDH) () is an enzyme that catalyzes the chemical reaction :L-histidinol + 2 NAD+ \rightleftharpoons L-histidine + 2 NADH + 2 H+ Thus, the two substrates of this enzyme are L-histidinol and NAD+, whereas its 3 products are L-histidine, NADH, and H+. This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-OH group of donor with NAD+ or NADP+ as acceptor. The systematic name of this enzyme class is L-histidinol:NAD+ oxidoreductase. This enzyme is also called L-histidinol dehydrogenase. Histidinol dehydrogenase catalyzes the terminal step in the biosynthesis of histidine in bacteria, fungi, and plants, the four-electron oxidation of L-histidinol to histidine. In 4-electron dehydrogenases, a single active site catalyses 2 separate oxidation steps: oxidation of the substrate alcohol to an intermediate aldehyde; and oxidation of the aldehyde to the product acid, in this case His. The reacti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
(R)-4-hydroxyphenyllactate Dehydrogenase
(R)-4-hydroxyphenyllactate dehydrogenase () is an enzyme that catalyzes a chemical reaction :(R)-3-(4-hydroxyphenyl)lactate + NAD(P)+ \rightleftharpoons 3-(4-hydroxyphenyl)pyruvate + NAD(P)H + H The 3 substrates of this enzyme are (R)-3-(4-hydroxyphenyl)lactate, NAD, and NADP, whereas its 4 products are 3-(4-hydroxyphenyl)pyruvate, NADH, NADPH, and H. This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-OH group of donor with NAD or NADP as acceptor. The systematic name of this enzyme class is (R)-3-(4-hydroxyphenyl)lactate:NAD(P) 2-oxidoreductase. Other names in common use include (R)-aromatic lactate dehydrogenase, and D-hydrogenase, D-aryllactate. This enzyme participates in tyrosine and phenylalanine Phenylalanine (symbol Phe or F) is an essential α-amino acid with the formula . It can be viewed as a benzyl group substituted for the methyl group of alanine, or a phenyl group in place of a terminal hydrogen of alanine. This essenti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UDP-glucose 6-dehydrogenase
UDP-glucose 6-dehydrogenase is a cytosolic enzyme that in humans is encoded by the ''UGDH'' gene. The protein encoded by this gene converts UDP-glucose to UDP-glucuronate and thereby participates in the biosynthesis of glycosaminoglycans such as hyaluronan, chondroitin sulfate, and heparan sulfate. These glycosylated compounds are common components of the extracellular matrix and likely play roles in signal transduction, cell migration, and cancer growth and metastasis. The expression of this gene is up-regulated by transforming growth factor beta and down-regulated by hypoxia. This enzyme participates in 4 metabolic pathways: pentose and glucuronate interconversions, ascorbate and aldarate metabolism, starch and sucrose metabolism, and nucleotide sugars metabolism. Loss of UGDH has recently been implicated in epileptic encephalopathy in humansHengel, H., Bosso-Lefèvre, C., Grady, G. et al. Loss-of-function mutations in UDP-Glucose 6-Dehydrogenase cause recessive developmental ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
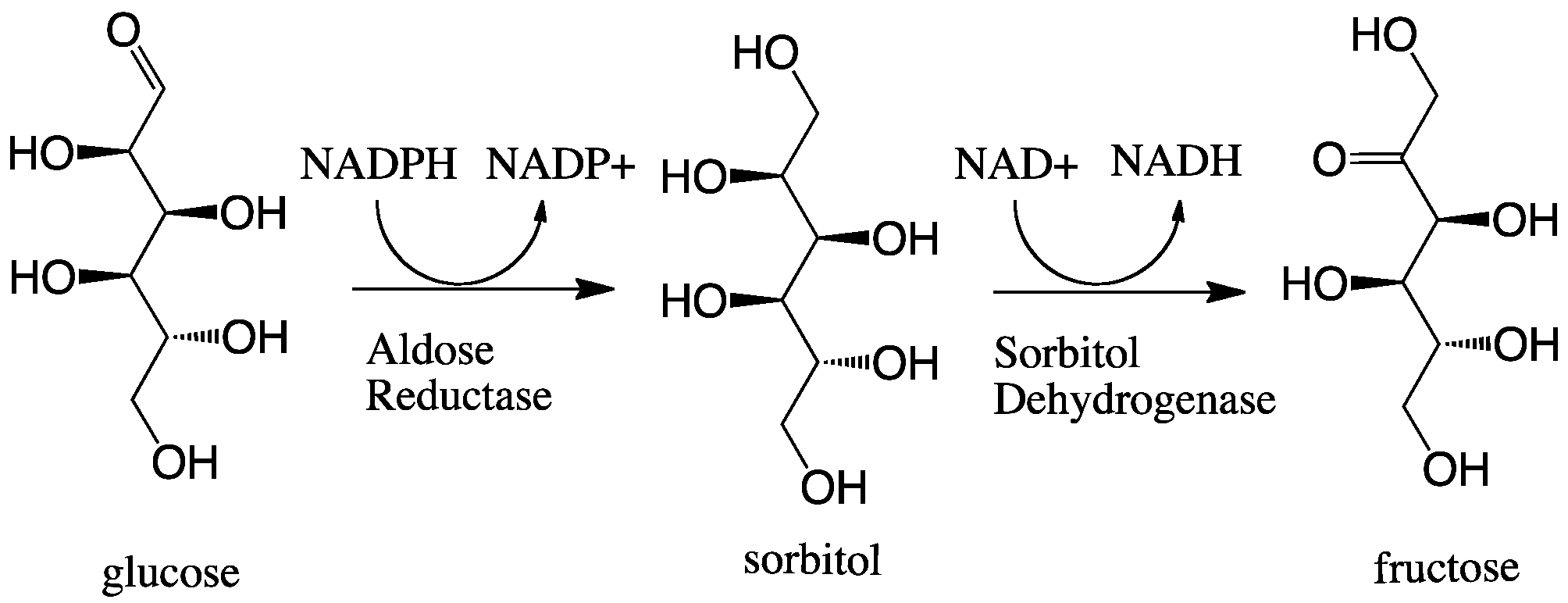
Aldose Reductase
In enzymology, aldose reductase (or aldehyde reductase) () is a cytosolic NADPH-dependent oxidoreductase that catalyzes the reduction of a variety of aldehydes and carbonyls, including monosaccharides. It is primarily known for catalyzing the reduction of glucose to sorbitol, the first step in polyol pathway of glucose metabolism. Reactions Aldose reductase catalyzes the NADPH-dependent conversion of glucose to sorbitol, the first step in polyol pathway of glucose metabolism. The second and last step in the pathway is catalyzed by sorbitol dehydrogenase, which catalyzes the NAD-linked oxidation of sorbitol to fructose. Thus, the polyol pathway results in conversion of glucose to fructose with stoichiometric utilization of NADPH and production of NADH. ;glucose + NADPH + H+ \rightleftharpoons sorbitol + NADP+ Galactose is also a substrate for the polyol pathway, but the corresponding keto sugar is not produced because sorbitol dehydrogenase is incapable of oxidizing galactitol. Ne ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
(+)-neomenthol Dehydrogenase
In enzymology, a (+)-neomenthol dehydrogenase () is an enzyme that catalyzes the chemical reaction :(+)-neomenthol + NADP \rightleftharpoons (−)-menthone + NADPH + H Thus, the two substrates of this enzyme are (+)-neomenthol and NADP, whereas its 3 products are (−)-menthone, NADPH, and H. This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-OH group of donor with NAD or NADP as acceptor. The systematic name of this enzyme class is (+)-neomenthol:NADP oxidoreductase. This enzyme is also called monoterpenoid dehydrogenase. This enzyme participates in monoterpenoid Monoterpenes are a class of terpenes that consist of two isoprene units and have the molecular formula C10H16. Monoterpenes may be linear (acyclic) or contain rings (monocyclic and bicyclic). Modified terpenes, such as those containing oxygen funct ... biosynthesis. References * EC 1.1.1 NADPH-dependent enzymes Enzymes of unknown structure {{1.1.1-enzyme-stu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
(-)-menthol Dehydrogenase
A (−)-menthol dehydrogenase () is an enzyme that catalyzes the chemical reaction :(−)-menthol + NADP \rightleftharpoons (−)-menthone + NADPH + H, i.e., catalyses the breakdown of menthol. Thus, the two substrates of this enzyme are (−)-menthol and NADP, whereas its 3 products are (−)-menthone, NADPH, and H. This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-OH group of donor with NAD or NADP as acceptor. The systematic name of this enzyme class is (−)-menthol:NADP oxidoreductase. This enzyme is also called monoterpenoid dehydrogenase. This enzyme participates in monoterpenoid Monoterpenes are a class of terpenes that consist of two isoprene units and have the molecular formula C10H16. Monoterpenes may be linear (acyclic) or contain rings (monocyclic and bicyclic). Modified terpenes, such as those containing oxygen funct ... biosynthesis. References * EC 1.1.1 NADPH-dependent enzymes Enzymes of unknown structure [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glucuronolactone Reductase
In enzymology, a glucuronolactone reductase () is an enzyme that catalyzes the chemical reaction :L-gulono-1,4-lactone + NADP+ \rightleftharpoons D-glucurono-3,6-lactone + NADPH + H+ Thus, the two substrates of this enzyme are L-gulono-1,4-lactone and NADP+, whereas its 3 products are D-glucurono-3,6-lactone, NADPH, and H+. This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-OH group of donor with NAD+ or NADP+ as acceptor. The systematic name of this enzyme class is L-gulono-1,4-lactone:NADP+ 1-oxidoreductase. Other names in common use include GRase, and gulonolactone dehydrogenase. This enzyme participates in ascorbate and aldarate metabolism Vitamin C (also known as ascorbic acid and ascorbate) is a water-soluble vitamin found in citrus and other fruits and vegetables, also sold as a dietary supplement and as a topical 'serum' ingredient to treat melasma (dark pigment spots) and .... References * EC 1.1.1 NADPH-depen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |