|
Ceramide Synthase 1
Ceramide synthase 1 also known as LAG1 longevity assurance homolog 1 is an enzyme that in humans is encoded by the ''CERS1'' gene. Function This gene encodes a member of the bone morphogenetic protein (BMP) family and the TGF-beta superfamily. This group of proteins is characterized by a polybasic proteolytic processing site that is cleaved to produce a mature protein containing seven conserved cysteine residues. Members of this family are regulators of cell growth and differentiation in both embryonic and adult tissues. Studies in yeast suggest that the encoded protein is involved in aging. This protein is transcribed from a monocistronic mRNA as well as a bicistronic mRNA, which also encodes growth differentiation factor 1. Ceramide synthase 1 (CerS1) is a ceramide synthase that catalyzes the synthesis of C18 ceramide in a fumonisin B1-independent manner, and is primarily expressed in the brain. It can also be found in low levels in skeletal muscle and the testis. With ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histochemistry And Cell Biology
''Histochemistry and Cell Biology'' is a peer-reviewed scientific journal in the field of molecular histology and cell biology, publishing original articles dealing with the localization and identification of molecular components, metabolic activities, and cell biological aspects of cells and tissues. The journal covers the development, application, and evaluation of methods and probes that can be used in the entire area of histochemistry and cell biology. The journal is published by Springer Science+Business Media and the official journal of the Society for Histochemistry. Earlier names of the journal are ''Histochemie'' and ''Histochemistry''. The editors-in-chief are Jürgen Roth ( University of Zurich), Takehiko Koji ( University of Nagasaki), Michael Schrader (University of Exeter) and Douglas J. Taatjes (University of Vermont The University of Vermont (UVM), officially the University of Vermont and State Agricultural College, is a Public university, public Land-grant un ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteasome
Proteasomes are protein complexes which degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. Enzymes that help such reactions are called proteases. Proteasomes are part of a major mechanism by which cells regulate the concentration of particular proteins and degrade misfolded proteins. Proteins are tagged for degradation with a small protein called ubiquitin. The tagging reaction is catalyzed by enzymes called ubiquitin ligases. Once a protein is tagged with a single ubiquitin molecule, this is a signal to other ligases to attach additional ubiquitin molecules. The result is a ''polyubiquitin chain'' that is bound by the proteasome, allowing it to degrade the tagged protein. The degradation process yields peptides of about seven to eight amino acids long, which can then be further degraded into shorter amino acid sequences and used in synthesizing new proteins. Proteasomes are found inside all eukaryotes and archaea, and in so ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitination
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A. The addition of ubiquitin to a substrate protein is called ubiquitylation (or, alternatively, ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide bond, cy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetic Tree
A phylogenetic tree (also phylogeny or evolutionary tree Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA.) is a branching diagram or a tree showing the evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical or genetic characteristics. All life on Earth is part of a single phylogenetic tree, indicating common ancestry. In a ''rooted'' phylogenetic tree, each node with descendants represents the inferred most recent common ancestor of those descendants, and the edge lengths in some trees may be interpreted as time estimates. Each node is called a taxonomic unit. Internal nodes are generally called hypothetical taxonomic units, as they cannot be directly observed. Trees are useful in fields of biology such as bioinformatics, systematics, and phylogenetics. ''Unrooted'' trees illustrate only the relatedness of the leaf nodes and do not require the ancestral root to b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homeobox
A homeobox is a DNA sequence, around 180 base pairs long, that regulates large-scale anatomical features in the early stages of embryonic development. For instance, mutations in a homeobox may change large-scale anatomical features of the full-grown organism. Homeoboxes are found within genes that are involved in the regulation of patterns of anatomical development (morphogenesis) in animal Animals are multicellular, eukaryotic organisms in the Kingdom (biology), biological kingdom Animalia. With few exceptions, animals Heterotroph, consume organic material, Cellular respiration#Aerobic respiration, breathe oxygen, are Motilit ...s, fungus, fungi, plants, and numerous single cell eukaryotes. Homeobox genes encode homeodomain protein products that are transcription factors sharing a characteristic protein fold structure that binds DNA to regulate expression of target genes. Homeodomain proteins regulate gene expression and cell differentiation during early embryonic dev ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Apoptosis
Apoptosis (from grc, ἀπόπτωσις, apóptōsis, 'falling off') is a form of programmed cell death that occurs in multicellular organisms. Biochemical events lead to characteristic cell changes (morphology) and death. These changes include blebbing, cell shrinkage, nuclear fragmentation, chromatin condensation, DNA fragmentation, and mRNA decay. The average adult human loses between 50 and 70 billion cells each day due to apoptosis. For an average human child between eight and fourteen years old, approximately twenty to thirty billion cells die per day. In contrast to necrosis, which is a form of traumatic cell death that results from acute cellular injury, apoptosis is a highly regulated and controlled process that confers advantages during an organism's life cycle. For example, the separation of fingers and toes in a developing human embryo occurs because cells between the digits undergo apoptosis. Unlike necrosis, apoptosis produces cell fragments called apoptotic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Granule Cells
A granule is a large particle or grain. It can refer to: * Granule (cell biology), any of several submicroscopic structures, some with explicable origins, others noted only as cell type-specific features of unknown function ** Azurophilic granule, a structure characteristic of the azurophil eukaryotic cell type ** Chromaffin granule, a structure characteristic of the chromophil eukaryotic cell type. * Astrophysics and geology: ** Granule (solar physics), a visible structure in the photosphere of the Sun arising from activity in the Sun's convective zone ** Martian spherules, spherical granules of material found on the surface of the planet Mars ** Granule (geology), a specified particle size of 2–4 millimetres (-1 to -2 on the φ scale) * Granule, in pharmaceutical terms, small particles gathered into a larger, permanent aggregate in which the original particles can still be identified * Granule (Oracle DBMS), a unit of contiguously allocated virtual memory * Granular synthesis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neurodegeneration
A neurodegenerative disease is caused by the progressive loss of structure or function of neurons, in the process known as neurodegeneration. Such neuronal damage may ultimately involve cell death. Neurodegenerative diseases include amyotrophic lateral sclerosis, multiple sclerosis, Parkinson's disease, Alzheimer's disease, Huntington's disease, multiple system atrophy, and prion diseases. Neurodegeneration can be found in the brain at many different levels of neuronal circuitry, ranging from molecular to systemic. Because there is no known way to reverse the progressive degeneration of neurons, these diseases are considered to be incurable; however research has shown that the two major contributing factors to neurodegeneration are oxidative stress and inflammation. Biomedical research has revealed many similarities between these diseases at the subcellular level, including atypical protein assemblies (like proteinopathy) and induced cell death. These similarities suggest that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Purkinje Neurons
Purkinje cells, or Purkinje neurons, are a class of GABAergic inhibitory neurons located in the cerebellum. They are named after their discoverer, Czech anatomist Jan Evangelista Purkyně, who characterized the cells in 1839. Structure These cells are some of the largest neurons in the human brain (Betz cells being the largest), with an intricately elaborate dendritic arbor, characterized by a large number of dendritic spines. Purkinje cells are found within the Purkinje layer in the cerebellum. Purkinje cells are aligned like dominos stacked one in front of the other. Their large dendritic arbors form nearly two-dimensional layers through which parallel fibers from the deeper-layers pass. These parallel fibers make relatively weaker excitatory (glutamatergic) synapses to spines in the Purkinje cell dendrite, whereas climbing fibers originating from the inferior olivary nucleus in the medulla provide very powerful excitatory input to the proximal dendrites and cell soma. Pa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
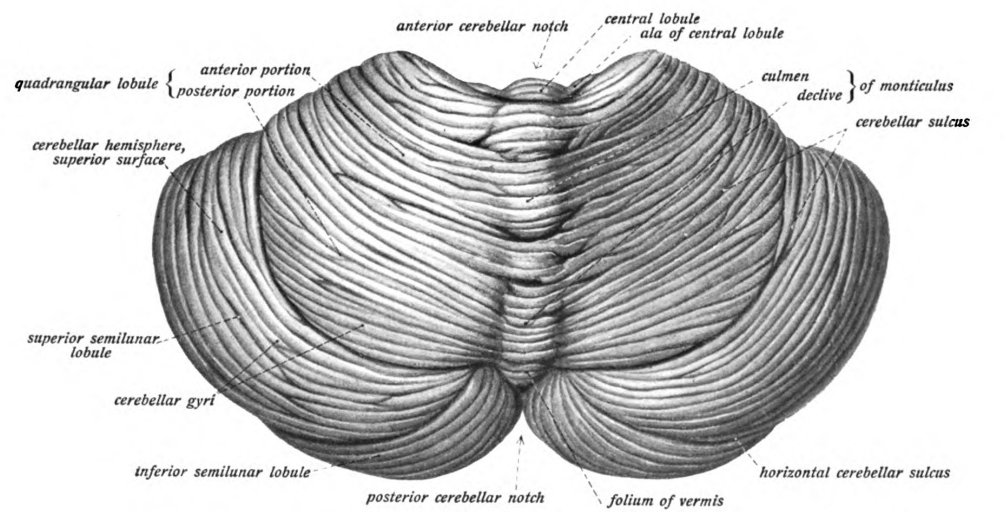
Cerebellum
The cerebellum (Latin for "little brain") is a major feature of the hindbrain of all vertebrates. Although usually smaller than the cerebrum, in some animals such as the mormyrid fishes it may be as large as or even larger. In humans, the cerebellum plays an important role in motor control. It may also be involved in some cognition, cognitive functions such as attention and language as well as emotion, emotional control such as regulating fear and pleasure responses, but its movement-related functions are the most solidly established. The human cerebellum does not initiate movement, but contributes to Motor coordination, coordination, precision, and accurate timing: it receives input from sensory systems of the spinal cord and from other parts of the brain, and integrates these inputs to fine-tune motor activity. Cerebellar damage produces disorders in Fine motor skill, fine movement, Equilibrioception, equilibrium, Human positions, posture, and motor learning in humans. Anatomica ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Atrophy
Atrophy is the partial or complete wasting away of a part of the body. Causes of atrophy include mutations (which can destroy the gene to build up the organ), poor nourishment, poor circulation, loss of hormonal support, loss of nerve supply to the target organ, excessive amount of apoptosis of cells, and disuse or lack of exercise or disease intrinsic to the tissue itself. In medical practice, hormonal and nerve inputs that maintain an organ or body part are said to have ''trophic'' effects. A diminished muscular trophic condition is designated as ''atrophy''. Atrophy is reduction in size of cell, organ or tissue, after attaining its normal mature growth. In contrast, hypoplasia is the reduction in the cellular numbers of an organ, or tissue that has not attained normal maturity. Atrophy is the general physiological process of reabsorption and breakdown of tissues, involving apoptosis. When it occurs as a result of disease or loss of trophic support because of other diseases ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |






.jpg)