|
Cell Cycle Regulated Methyltransferase
CcrM (or M.CcrMI) is an orphan DNA methyltransferase, that is involved in controlling gene expression in most Alphaproteobacteria. This enzyme modifies DNA by catalyzing the transference of a methyl group from the S-adenosyl-L methionine substrate to the N6 position of an adenine base in the sequence 5'-GANTC-3' with high specificity. In some lineages such as SAR11, the homologous enzymes possess 5'-GAWTC-3' specificity. In ''Caulobacter crescentus'' Ccrm is produced at the end of the replication cycle when Ccrm recognition sites are hemimethylated, rapidly methylating the DNA. CcrM is essential in other Alphaproteobacteria but its role is not yet determined. CcrM is a highly specific methyltransferase with a novel DNA recognition mechanism. CcrM role in cell cycle regulation Methylations are epigenetic modification that, in eukaryotes, regulates processes as cell differentiation, and embryogenesis, while in prokaryotes can have a role in self recognition, protecting the DNA from ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Caulobacter Crescentus CcrM
''Caulobacter'' is a genus of Gram-negative bacteria in the class Alphaproteobacteria. Its best-known member is ''Caulobacter crescentus'', an organism ubiquitous in freshwater lakes and rivers; many members of the genus are specialized to Trophic state index#Oligotrophic, oligotrophic environments. Interactions with other organisms Pathogenicity Although ''Caulobacter'' is not commonly appreciated as a cause of human diseases, ''Caulobacter'' isolates have been implicated in a number of cases of recurrent peritonitis in peritoneal dialysis patients. One study has identified the species ''C. crescentus'' and ''C. mirare'' as the cause of a disease of the moth ''Galleria mellonella''; the absence of identified distinct virulence factors in ''C. mirare'' may suggest that other ''Caulobacter'' species have pathogenic potential. References External links * https://microbewiki.kenyon.edu/index.php/Caulobacter * https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Undef& ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
S Phase
S phase (Synthesis Phase) is the phase of the cell cycle in which DNA is replicated, occurring between G1 phase and G2 phase. Since accurate duplication of the genome is critical to successful cell division, the processes that occur during S-phase are tightly regulated and widely conserved. Regulation Entry into S-phase is controlled by the G1 restriction point (R), which commits cells to the remainder of the cell-cycle if there is adequate nutrients and growth signaling. This transition is essentially irreversible; after passing the restriction point, the cell will progress through S-phase even if environmental conditions become unfavorable. Accordingly, entry into S-phase is controlled by molecular pathways that facilitate a rapid, unidirectional shift in cell state. In yeast, for instance, cell growth induces accumulation of Cln3 cyclin, which complexes with the cyclin dependent kinase CDK2. The Cln3-CDK2 complex promotes transcription of S-phase genes by inactivating ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gammaproteobacteria
Gammaproteobacteria is a class of bacteria in the phylum Pseudomonadota (synonym Proteobacteria). It contains about 250 genera, which makes it the most genera-rich taxon of the Prokaryotes. Several medically, ecologically, and scientifically important groups of bacteria belong to this class. It is composed by all Gram-negative microbes and is the most phylogenetically and physiologically diverse class of Proteobacteria. These microorganisms can live in several terrestrial and marine environments, in which they play various important roles, including ''extreme environments'' such as hydrothermal vents. They generally have different shapes - rods, curved rods, cocci, spirilla, and filaments and include free living bacteria, biofilm formers, commensals and symbionts, some also have the distinctive trait of being bioluminescent. Metabolisms found in the different genera are very different; there are both aerobic and anaerobic (obligate or facultative) species, chemolithoautotrophic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Campylobacterota
Campylobacterota are a phylum of bacteria. All species of this phylum are Gram-negative. The Campylobacterota consist of few known genera, mainly the curved to spirilloid ''Wolinella'' spp., ''Helicobacter'' spp., and '' Campylobacter'' spp. Most of the known species inhabit the digestive tracts of animals and serve as symbionts (''Wolinella'' spp. in cattle) or pathogens (''Helicobacter'' spp. in the stomach, ''Campylobacter'' spp. in the duodenum). Many Campylobacterota are motile with flagella. Numerous environmental sequences and isolates of Campylobacterota have also been recovered from hydrothermal vents and cold seep habitats. Examples of isolates include ''Sulfurimonas autotrophica'', ''Sulfurimonas paralvinellae'', ''Sulfurovum lithotrophicum'' and ''Nautilia profundicola''. A member of the phylum Campylobacterota occurs as an endosymbiont in the large gills of the deepwater sea snail ''Alviniconcha hessleri''. The Campylobacterota found at deep-sea hydrothermal vents ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Magnetococcales
The Magnetococcales were an order of Alphaproteobacteria, but now the mitochondria are considered as sister to the alphaproteobactera, together forming the sister the marineproteo1 group, together forming the sister to Magnetococcidae. See also * List of bacterial orders * List of bacteria genera This article lists the genera of the bacteria. The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN) and National Center for Biotechnology Information (NCBI). However many taxonomic names are ... References Alphaproteobacteria Bacteria orders {{alphaproteobacteria-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rickettsiales
The Rickettsiales, informally called rickettsias, are an order of small Alphaproteobacteria. They are obligate intracellular parasites, and some are notable pathogens, including ''Rickettsia'', which causes a variety of diseases in humans, and ''Ehrlichia'', which causes diseases in livestock. Another genus of well-known Rickettsiales is the ''Wolbachia'', which infect about two-thirds of all arthropods and nearly all filarial nematodes. Genetic studies support the endosymbiotic theory according to which mitochondria and related organelles developed from members of this group.Thomas S. (2016). Rickettsiales:Biology, Molecular Biology, Epidemiology, and Vaccine Development. pp.529. Springer• The Rickettsiales are difficult to culture, as they rely on living eukaryotic host cells for their survival. Rickettsiales phylogeny The Rickettsiales further consist of three known families, the Rickettsiaceae, the Midichloriaceae, and the Ehrlichiaceae. Most studies also support the in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lon Protease Family
In molecular biology, the Lon protease family is a family of enzymes that break peptide bonds in proteins resulting in smaller peptides or amino acids. They are found in archaea, bacteria and eukaryotes. Lon proteases are ATP-dependent serine peptidases belonging to the MEROPS peptidase family S16 (Lon protease family, clan SJ). In the eukaryotes the majority of the Lon proteases are located in the mitochondrial matrix. In yeast, the Lon protease PIM1 is located in the mitochondrial matrix. It is required for mitochondrial function, it is constitutively expressed but is increased after thermal stress, suggesting that PIM1 may play a role in the heat shock The heat shock response (HSR) is a cell stress response that increases the number of molecular chaperones to combat the negative effects on proteins caused by stressors such as increased temperatures, oxidative stress, and heavy metals. In a normal ... response. Lon proteases have two specific subfamilies: LonA and LonB, diffe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
G1 Phase
The G1 phase, gap 1 phase, or growth 1 phase, is the first of four phases of the cell cycle that takes place in eukaryotic cell division. In this part of interphase, the cell synthesizes mRNA and proteins in preparation for subsequent steps leading to mitosis. G1 phase ends when the cell moves into the S phase of interphase. Around 30 to 40 percent of cell cycle time is spent in the G1 phase. Overview G1 phase together with the S phase and G2 phase comprise the long growth period of the cell cycle cell division called interphase that takes place before cell division in mitosis (M phase). During G1 phase, the cell grows in size and synthesizes mRNA and protein that are required for DNA synthesis. Once the required proteins and growth are complete, the cell enters the next phase of the cell cycle, S phase. The duration of each phase, including the G1 phase, is different in many different types of cells. In human somatic cells, the cell cycle lasts about 10 hours, and the G1 H ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dam Methylase
DNA adenine methylase, (Dam methylase) (also site-specific DNA-methyltransferase (adenine-specific), , ''modification methylase'', ''restriction-modification system'') is an enzyme that adds a methyl group to the adenine of the sequence 5'-GATC-3' in newly synthesized DNA. Immediately after DNA synthesis, the daughter strand remains unmethylated for a short time. It is an orphan methyltransferase that is not part of a restriction-modification system and regulates gene expression. This enzyme catalyses the following chemical reaction : S-adenosyl-L-methionine + DNA adenine \rightleftharpoons S-adenosyl-L-homocysteine + DNA 6-methylaminopurine This is a large group of enzymes unique to prokaryotes and bacteriophages. The ''E. coli'' DNA adenine methyltransferase enzyme (Dam), is widely used for the chromatin profiling technique, DamID. In which the Dam is fused to a DNA-binding protein of interest and expressed as a transgene in a genetically tractable model organism to identif ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Methyltransferase
In biochemistry, the DNA methyltransferase (DNA MTase, DNMT) family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl methionine (SAM) as the methyl donor. Classification Substrate MTases can be divided into three different groups on the basis of the chemical reactions they catalyze: * m6A - those that generate N6-methyladenine * m4C - those that generate N4-methylcytosine * m5C - those that generate C5-methylcytosine m6A and m4C methyltransferases are found primarily in prokaryotes (although recent evidence has suggested that m6A is abundant in eukaryotes). m5C methyltransfereases are found in some lower eukaryotes, in most higher plants, and in animals beginning with the echinoderms. The m6A methyltransferases (N-6 adenine-specific DNA methylase) (A-Mtase) are enzymes that specifically methylate the amino group at the C-6 position of adenine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
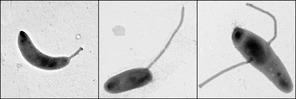
Caulobacter Crescentus
''Caulobacter crescentus'' is a Gram-negative, oligotrophic bacterium widely distributed in fresh water lakes and streams. The taxon is more properly known as ''Caulobacter vibrioides'' (Henrici and Johnson 1935). ''C. crescentus'' is an important model organism for studying the regulation of the cell cycle, asymmetric cell division, and cellular differentiation. ''Caulobacter'' daughter cells have two very different forms. One daughter is a mobile "swarmer" cell that has a single flagellum at one cell pole that provides swimming motility for chemotaxis. The other daughter, called the "stalked" cell, has a tubular stalk structure protruding from one pole that has an adhesive holdfast material on its end, with which the stalked cell can adhere to surfaces. Swarmer cells differentiate into stalked cells after a short period of motility. Chromosome replication and cell division only occurs in the stalked cell stage. ''C. crescentus'' derives its name from its crescent shape, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SAR11
The Pelagibacterales are an order in the Alphaproteobacteria composed of free-living marine bacteria that make up roughly one in three cells at the ocean's surface. Overall, members of the ''Pelagibacterales'' are estimated to make up between a quarter and a half of all prokaryotic cells in the ocean. Initially, this taxon was known solely by metagenomic data and was known as the SAR11 clade. It was first placed in the Rickettsiales, but was later raised to the rank of order, and then placed as sister order to the Rickettsiales in the subclass Rickettsidae. It includes the highly abundant marine species '' Pelagibacter ubique''. Bacteria in this order are unusually small. Due to their small genome size and limited metabolic function, ''Pelagibacterales'' have become a model organism for 'streamlining theory'. ''P. ubique'' and related species are oligotrophs (scavengers) and feed on dissolved organic carbon and nitrogen. They are unable to fix carbon or nitrogen, but can p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |