|
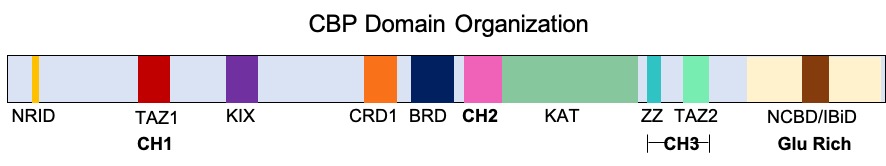
CREB-binding Protein
CREB-binding protein, also known as CREBBP or CBP or KAT3A, (where CREB is cAMP response element-binding protein) is a coactivator encoded by the ''CREBBP'' gene in humans, located on chromosome 16p13.3. CBP has intrinsic acetyltransferase functions; it is able to add acetyl groups to both transcription factors as well as histone lysines, the latter of which has been shown to alter chromatin structure making genes more accessible for transcription. This relatively unique acetyltransferase activity is also seen in another transcription enzyme, EP300 (p300). Together, they are known as the p300-CBP coactivator family and are known to associate with more than 16,000 genes in humans; however, while these proteins share many structural features, emerging evidence suggests that these two co-activators may promote transcription of genes with different biological functions. For example, CBP alone has been implicated in a wide variety of pathophysiologies including colorectal cancer as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CREB
CREB-TF (CREB, cAMP response element-binding protein) is a cellular transcription factor. It binds to certain DNA sequences called cAMP response elements (CRE), thereby increasing or decreasing the transcription of the genes. CREB was first described in 1987 as a cAMP-responsive transcription factor regulating the somatostatin gene. Genes whose transcription is regulated by CREB include: '' c-fos'', BDNF, tyrosine hydroxylase, numerous neuropeptides (such as somatostatin, enkephalin, VGF, corticotropin-releasing hormone), and genes involved in the mammalian circadian clock (PER1, PER2). CREB is closely related in structure and function to CREM ( cAMP response element modulator) and ATF-1 ( activating transcription factor-1) proteins. CREB proteins are expressed in many animals, including humans. CREB has a well-documented role in neuronal plasticity and long-term memory formation in the brain and has been shown to be integral in the formation of spatial memory. CREB down ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Depression (mood)
Depression is a mental state of low Mood (psychology), mood and aversion to activity. It affects about 3.5% of the global population, or about 280 million people worldwide, as of 2020. Depression affects a person's thoughts, behavior, feelings, and subjective well-being, sense of well-being. The pleasure or joy that a person gets from certain experiences is reduced, and the afflicted person often experiences a loss of motivation or interest in those activities. People with depression may experience sadness, feelings of dejection or hopelessness, difficulty in thinking and concentration, or a significant change in appetite or time spent sleeping; Suicidal ideation, suicidal thoughts can also be experienced. Depression can have multiple, sometimes overlapping, origins. Depression can be a symptom of some mood disorders, some of which are also commonly called ''depression'', such as major depressive disorder, bipolar disorder and dysthymia. Additionally, depression can be a norm ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Receptor Coactivator 1
The nuclear receptor coactivator 1 (NCOA1), also called steroid receptor coactivator-1 (SRC-1), is a transcriptional coregulatory protein that contains several nuclear receptor–interacting domains and possesses intrinsic histone acetyltransferase activity. It is encoded by the gene ''NCOA1''. NCOA1 is recruited to DNA promoter sites by ligand-activated nuclear receptors. NCOA1, in turn, acylates histones, which makes downstream DNA more accessible to transcription. Hence, NCOA1 assists nuclear receptors in the upregulation of DNA expression as a coactivator. Interactions Nuclear receptor coactivator 1 possesses a basic helix-loop-helix (bHLH) domain and has been shown to interact with: * Androgen receptor, * C-Fos, * C-jun, * CIITA, * CREB-binding protein, * Cyclin D1, * Estrogen receptor alpha, * Glucocorticoid receptor, * NFKB1, * PCAF, * PPARGC1A, * Peroxisome proliferator-activated receptor alpha, * SNW1, * STAT3 Signal transducer and activator ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MyoD
MyoD, also known as myoblast determination protein 1, is a protein in animals that plays a major role in regulating muscle differentiation. MyoD, which was discovered in the laboratory of Harold M. Weintraub, belongs to a family of proteins known as myogenic regulatory factors (MRFs). These bHLH (basic helix loop helix) transcription factors act sequentially in myogenic differentiation. Vertebrate MRF family members include MyoD1, Myf5, myogenin, and MRF4 (Myf6). In non-vertebrate animals, a single MyoD protein is typically found. MyoD is one of the earliest markers of myogenic commitment. MyoD is expressed at extremely low and essentially undetectable levels in quiescent satellite cells, but expression of MyoD is activated in response to exercise or muscle tissue damage. The effect of MyoD on satellite cells is dose-dependent; high MyoD expression represses cell renewal, promotes terminal differentiation and can induce apoptosis. Although MyoD marks myoblast commitment, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GATA1
GATA-binding factor 1 or GATA-1 (also termed Erythroid transcription factor) is the founding member of the GATA family of transcription factors. This protein is widely expressed throughout vertebrate species. In humans and mice, it is encoded by the ''GATA1'' and ''Gata1'' genes, respectively. These genes are located on the X chromosome in both species. GATA1 regulates the expression (i.e. formation of the genes' products) of an ensemble of genes that mediate the development of red blood cells and platelets. Its critical roles in red blood cell formation include promoting the maturation of precursor cells, e.g. erythroblasts, to red blood cells and stimulating these cells to erect their cytoskeleton and biosynthesize their oxygen-carrying components viz., hemoglobin and heme. GATA1 plays a similarly critical role in the maturation of blood platelets from megakaryoblasts, promegakaryocytes, and megakaryocytes; the latter cells then shed membrane-enclosed fragments of their c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphorylation
In biochemistry, phosphorylation is described as the "transfer of a phosphate group" from a donor to an acceptor. A common phosphorylating agent (phosphate donor) is ATP and a common family of acceptor are alcohols: : This equation can be written in several ways that are nearly equivalent that describe the behaviors of various protonated states of ATP, ADP, and the phosphorylated product. As is clear from the equation, a phosphate group per se is not transferred, but a phosphoryl group (PO3-). Phosphoryl is an electrophile. This process and its inverse, dephosphorylation, are common in biology. Text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. Protein phosphorylation often activates (or deactivates) many enzymes. During respiration Phosphorylation is essential to the processes of both anaerobic and aerobic respiration, which involve the production of adenosine triphosphate (ATP), the "high-energy" exc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
STAT3
Signal transducer and activator of transcription 3 (STAT3) is a transcription factor which in humans is encoded by the ''STAT3'' gene. It is a member of the STAT protein family. Function STAT3 is a member of the STAT protein family. In response to cytokines and growth factors, STAT3 is phosphorylated by receptor-associated Janus kinases (JAK), forms homo- or heterodimers, and translocates to the cell nucleus where it acts as a transcription activators, transcription activator. Specifically, STAT3 becomes activated after phosphorylation of tyrosine 705 in response to such ligands as interferons, epidermal growth factor (EGF), Interleukin 5, interleukin (IL-)5 and Interleukin 6, IL-6. Additionally, activation of STAT3 may occur via phosphorylation of serine 727 by mitogen-activated protein kinases (MAPK) and through Src (gene), c-src non-receptor tyrosine kinase. STAT3 mediates the expression of a variety of genes in response to cell stimuli, and thus plays a key role in many cel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MYB (gene)
Myb genes are part of a large gene family of transcription factors found in animals and plants. In humans, it includes Myb proto-oncogene like 1 and Myb-related protein B in addition to MYB proper. Members of the extended SANT/Myb family also include the SANT domain and other similar all-helical homeobox-like domains. Function Viral The Myb gene family is named after the eponymous gene in Alpharetrovirus, Avian myeloblastosis virus. The viral Myb (v-Myb, ) recognizes the sequence 5'-YAACKG-3'. It causes myeloblastosis (myeloid leukemia) in chickens. Compared to the normal animal cellular Myb (c-myb), v-myb contains deletions in the C-terminal regulatory domain, leading to aberrant activation of other oncogenes. Animals Myb proto-oncogene protein is a member of the MYB (myeloblastosis) family of transcription factors. The protein contains three domains, an N-terminus, N-terminal DNA-binding domain, a central transcriptional activation domain and a C-terminal domain involved ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
KMT2A
Histone-lysine ''N''-methyltransferase 2A, also known as acute lymphoblastic leukemia 1 (ALL-1), myeloid/lymphoid or mixed-lineage leukemia 1 (MLL1), or zinc finger protein HRX (HRX), is an enzyme that in humans is encoded by the ''KMT2A'' gene. MLL1 is a histone methyltransferase deemed a positive global regulator of gene transcription. This protein belongs to the group of histone-modifying enzymes comprising transactivation domain 9aaTAD; ; and is involved in the epigenetic maintenance of transcriptional memory. Its role as an epigenetic regulator of neuronal function is an ongoing area of research. Function Transcriptional regulation KMT2A gene encodes a transcriptional coactivator that plays an essential role in regulating gene expression during early development and hematopoiesis. The encoded protein contains multiple conserved functional domains. One of these domains, the SET domain, is responsible for its histone H3 lysine 4 (H3K4) methyltransferase activity whi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
KIX Domain
In biochemistry, the KIX domain (kinase-inducible domain (KID) interacting domain) or CREB binding domain is a protein domain of the eukaryotic transcriptional coactivators CBP and P300. It serves as a docking site for the formation of heterodimers between the coactivator and specific transcription factors. Structurally, the KIX domain is a globular domain consisting of three α-helices and two short 310-helices. The KIX domain was originally discovered in 1996 as the specific and minimal region in CBP that binds and interacts with phosphorylated CREB to activate transcription. It was thus first termed CREB-binding domain. However, when it was later discovered that it also binds many other proteins, the more general name KIX domain became favoured. The KIX domain contains two separate binding sites: the "c-Myb site", named after the oncoprotein c-Myb, and the "MLL site", named after the proto-oncogene MLL (Mixed Lineage Leukemia, KMT2A). The paralogous coactivators CBP ( CREB ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Primary Structure
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthesis is most commonly performed by ribosomes in cells. Peptides can also be synthesized in the laboratory. Protein primary structures can be directly sequenced, or inferred from DNA sequences. Formation Biological Amino acids are polymerised via peptide bonds to form a long backbone, with the different amino acid side chains protruding along it. In biological systems, proteins are produced during translation by a cell's ribosomes. Some organisms can also make short peptides by non-ribosomal peptide synthesis, which often use amino acids other than the encoded 22, and may be cyclised, modified and cross-linked. Chemical Peptides can be synthesised chemically via a range of laboratory methods. Chemical methods typically synthe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |