|
Bacteriophage MS2
Bacteriophage MS2 (''Emesvirus zinderi''), commonly called MS2, is an icosahedral, positive-sense single-stranded RNA virus that infects the bacterium ''Escherichia coli'' and other members of the Enterobacteriaceae. MS2 is a member of a family of closely related bacterial viruses that includes bacteriophage f2, bacteriophage Qβ, R17, and GA. History In 1961, MS2 was isolated by Alvin John Clark and recognized as an RNA-containing phage very similar to bacteriophage f2. In 1976, the MS2 genome was the first genome to be completely sequenced. This was accomplished by Walter Fiers and his team, building upon their earlier milestone in 1972 of the first gene to be completely sequenced, the MS2 coat protein. These sequences were determined at the RNA level, whereas the next landmark achievement, the sequence of the bacteriophage ΦX174 genome in 1977, was determined using DNA. The first effort at a statistical analysis of the MS2 genome was a search for patterns in the nucleoti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Viral Capsid
A capsid is the protein shell of a virus, enclosing its genetic material. It consists of several oligomeric (repeating) structural subunits made of protein called protomers. The observable 3-dimensional morphological subunits, which may or may not correspond to individual proteins, are called capsomeres. The proteins making up the capsid are called capsid proteins or viral coat proteins (VCP). The capsid and inner genome is called the nucleocapsid. Capsids are broadly classified according to their structure. The majority of the viruses have capsids with either helical or icosahedral structure. Some viruses, such as bacteriophages, have developed more complicated structures due to constraints of elasticity and electrostatics. The icosahedral shape, which has 20 equilateral triangular faces, approximates a sphere, while the helical shape resembles the shape of a spring, taking the space of a cylinder but not being a cylinder itself. The capsid faces may consist of one or m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
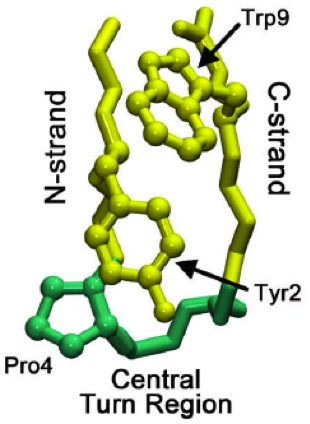
Beta Hairpin
The beta hairpin (sometimes also called beta-ribbon or beta-beta unit) is a simple protein structural motif involving two beta strands that look like a hairpin. The motif consists of two strands that are adjacent in primary structure, oriented in an antiparallel direction (the N-terminus of one sheet is adjacent to the C-terminus of the next), and linked by a short loop of two to five amino acids. Beta hairpins can occur in isolation or as part of a series of hydrogen bonded strands that collectively comprise a beta sheet. Researchers such as Francisco Blanco ''et al.'' have used protein NMR to show that beta-hairpins can be formed from isolated short peptides in aqueous solution, suggesting that hairpins could form nucleation sites for protein folding. Classification Beta hairpins were originally categorized solely by the number of amino acid residues in their loop sequences, such that they were named one-residue, two-residue, etc. This system, however, is somewhat ambiguous ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pilus
A pilus (Latin for 'hair'; plural: ''pili'') is a hair-like appendage found on the surface of many bacteria and archaea. The terms ''pilus'' and '' fimbria'' (Latin for 'fringe'; plural: ''fimbriae'') can be used interchangeably, although some researchers reserve the term ''pilus'' for the appendage required for bacterial conjugation. All conjugative pili are primarily composed of pilin – fibrous proteins, which are oligomeric. Dozens of these structures can exist on the bacterial and archaeal surface. Some bacteria, viruses or bacteriophages attach to receptors on pili at the start of their reproductive cycle. Pili are antigenic. They are also fragile and constantly replaced, sometimes with pili of different composition, resulting in altered antigenicity. Specific host responses to old pili structures are not effective on the new structure. Recombination genes of pili code for variable (V) and constant (C) regions of the pili (similar to immunoglobulin diversity). As the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacterial Conjugation
Bacterial conjugation is the transfer of genetic material between bacterial cells by direct cell-to-cell contact or by a bridge-like connection between two cells. This takes place through a pilus. It is a parasexual mode of reproduction in bacteria. It is a mechanism of horizontal gene transfer as are transformation and transduction although these two other mechanisms do not involve cell-to-cell contact. Classical ''E. coli'' bacterial conjugation is often regarded as the bacterial equivalent of sexual reproduction or mating since it involves the exchange of genetic material. However, it is not sexual reproduction, since no exchange of gamete occurs, and indeed no generation of a new organism: instead an existing organism is transformed. During classical ''E. coli'' conjugation the ''donor'' cell provides a conjugative or mobilizable genetic element that is most often a plasmid or transposon. Most conjugative plasmids have systems ensuring that the ''recipient'' cell does not al ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria; however, plasmids are sometimes present in archaea and eukaryotic organisms. In nature, plasmids often carry genes that benefit the survival of the organism and confer selective advantage such as antibiotic resistance. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain only additional genes that may be useful in certain situations or conditions. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the inter ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fertility Factor (bacteria)
The fertility factor (first named F by one of its discoverers Esther Lederberg; also called the sex factor in '' E. coli'' or the F sex factor; also called F-plasmid) allows genes to be transferred from one bacterium carrying the factor to another bacterium lacking the factor by conjugation. The F factor was the first plasmid to be discovered. Unlike other plasmids, F factor is constitutive for transfer proteins due to a mutation in the gene ''finO''. The F plasmid belongs to a class of conjugative plasmids that control sexual functions of bacteria with a fertility inhibition (Fin) system. Discovery Esther M. Lederberg and Luigi L. Cavalli-Sforza discovered "F," subsequently publishing with Joshua Lederberg. Once her results were announced, two other labs joined the studies. "This was not a simultaneous independent discovery of F (I names as Fertility Factor until it was understood.) We wrote to Hayes, Jacob, & Wollman who then proceeded with their studies." The discovery of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid Secondary Structure
Nucleic acid secondary structure is the basepairing interactions within a single nucleic acid polymer or between two polymers. It can be represented as a list of bases which are paired in a nucleic acid molecule. The secondary structures of biological DNAs and RNAs tend to be different: biological DNA mostly exists as fully base paired double helices, while biological RNA is single stranded and often forms complex and intricate base-pairing interactions due to its increased ability to form hydrogen bonds stemming from the extra hydroxyl group in the ribose sugar. In a non-biological context, secondary structure is a vital consideration in the nucleic acid design of nucleic acid structures for DNA nanotechnology and DNA computing, since the pattern of basepairing ultimately determines the overall structure of the molecules. Fundamental concepts Base pairing In molecular biology, two nucleotides on opposite complementary DNA or RNA strands that are connected via hydrogen b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Translation (biology)
In molecular biology and genetics, translation is the process in which ribosomes in the cytoplasm or endoplasmic reticulum synthesize proteins after the process of transcription (biology), transcription of DNA to RNA in the cell's nucleus (cell), nucleus. The entire process is called gene expression. In translation, mRNA, messenger RNA (mRNA) is decoded in a ribosome, outside the nucleus, to produce a specific amino acid chain, or polypeptide. The polypeptide later protein folding, folds into an Activation energy, active protein and performs its functions in the Cell (biology), cell. The ribosome facilitates decoding by inducing the binding of Base pair, complementary tRNA anticodon sequences to mRNA codons. The tRNAs carry specific amino acids that are chained together into a polypeptide as the mRNA passes through and is "read" by the ribosome. Translation proceeds in three phases: # Initiation: The ribosome assembles around the target mRNA. The first tRNA is attached a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication. The process of gene expression is used by all known life—eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and utilized by viruses—to generate the macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phenotype, '' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein. mRNA is created during the process of transcription, where an enzyme (RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and, utilising amino acids carried by transfer RNA (tRNA), the ribosome creates the protein. This process is known as translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genetic information in a biological system. As in DNA, genetic inf ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Overlapping Gene
An overlapping gene (or OLG) is a gene whose expressible Nucleic acid sequence, nucleotide sequence partially overlaps with the expressible nucleotide sequence of another gene. In this way, a nucleotide sequence may make a contribution to the function of one or more gene products. Overlapping genes are present and a fundamental feature of both Cell (biology), cellular and Virus, viral genomes. The current definition of an overlapping gene varies significantly between eukaryotes, prokaryotes, and viruses. In prokaryotes and viruses overlap must be between Coding region, coding sequences but not Messenger RNA, mRNA transcripts, and is defined when these coding sequences share a nucleotide on either the same or opposite strands. In eukaryotes, gene overlap is almost always defined as mRNA transcript overlap. Specifically, a gene overlap in eukaryotes is defined when at least one nucleotide is shared between the boundaries of the primary mRNA transcripts of two or more genes, such that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Replicase
RNA-dependent RNA polymerase (RdRp) or RNA replicase is an enzyme that catalyzes the replication of RNA from an RNA template. Specifically, it catalyzes synthesis of the RNA strand complementary to a given RNA template. This is in contrast to typical DNA-dependent RNA polymerases, which all organisms use to catalyze the transcription of RNA from a DNA template. RdRp is an essential protein encoded in the genomes of most RNA-containing viruses with no DNA stage including SARS-CoV-2. Some eukaryotes also contain RdRps, which are involved in RNA interference and differ structurally from viral RdRps. History Viral RdRps were discovered in the early 1960s from studies on mengovirus and polio virus when it was observed that these viruses were not sensitive to actinomycin D, a drug that inhibits cellular DNA-directed RNA synthesis. This lack of sensitivity suggested that there is a virus-specific enzyme that could copy RNA from an RNA template and not from a DNA template. D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |

.gif)