|
Biomolecular Structure
Biomolecular structure is the intricate folded, three-dimensional shape that is formed by a molecule of protein, DNA, or RNA, and that is important to its function. The structure of these molecules may be considered at any of several length scales ranging from the level of individual atoms to the relationships among entire protein subunits. This useful distinction among scales is often expressed as a decomposition of molecular structure into four levels: primary, secondary, tertiary, and quaternary. The scaffold for this multiscale organization of the molecule arises at the secondary level, where the fundamental structural elements are the molecule's various hydrogen bonds. This leads to several recognizable ''domains'' of protein structure and nucleic acid structure, including such secondary-structure features as alpha helixes and beta sheets for proteins, and hairpin loops, bulges, and internal loops for nucleic acids. The terms ''primary'', ''secondary'', ''tertiary'', and ''q ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecule
A molecule is a group of two or more atoms that are held together by Force, attractive forces known as chemical bonds; depending on context, the term may or may not include ions that satisfy this criterion. In quantum physics, organic chemistry, and biochemistry, the distinction from ions is dropped and ''molecule'' is often used when referring to polyatomic ions. A molecule may be homonuclear, that is, it consists of atoms of one chemical element, e.g. two atoms in the oxygen molecule (O2); or it may be heteronuclear, a chemical compound composed of more than one element, e.g. water (molecule), water (two hydrogen atoms and one oxygen atom; H2O). In the kinetic theory of gases, the term ''molecule'' is often used for any gaseous particle regardless of its composition. This relaxes the requirement that a molecule contains two or more atoms, since the noble gases are individual atoms. Atoms and complexes connected by non-covalent interactions, such as hydrogen bonds or ionic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Primary Structure
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthesis is most commonly performed by ribosomes in cells. Peptides can also be synthesized in the laboratory. Protein primary structures can be directly sequenced, or inferred from DNA sequences. Formation Biological Amino acids are polymerised via peptide bonds to form a long backbone, with the different amino acid side chains protruding along it. In biological systems, proteins are produced during translation by a cell's ribosomes. Some organisms can also make short peptides by non-ribosomal peptide synthesis, which often use amino acids other than the encoded 22, and may be cyclised, modified and cross-linked. Chemical Peptides can be synthesised chemically via a range of laboratory methods. Chemical methods typically synthe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
U12 Minor Spliceosomal RNA
U12 minor spliceosomal RNA is formed from U12 small nuclear ( snRNA), together with U4atac/ U6atac, U5, and U11 snRNAs and associated proteins, forms a spliceosome that cleaves a divergent class of low-abundance pre-mRNA introns. Although the U12 sequence is very divergent from that of U2, the two are functionally analogous. Structure The predicted secondary structure of U12 RNA is published,. However, the alternative single hairpin in the 3' end shown here seems to better match the alignment of divergent ''Drosophila melanogaster'' and ''Arabidopsis thaliana'' sequences. The sequences U12 introns that are spliced out are collected in a biological database Biological databases are libraries of biological sciences, collected from scientific experiments, published literature, high-throughput experiment technology, and computational analysis. They contain information from research areas including geno ... called the U12 intron database. References External links * ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
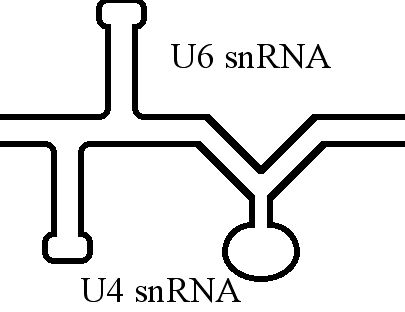
U6 Spliceosomal RNA
U6 snRNA is the non-coding small nuclear RNA (snRNA) component of U6 snRNP (''small nuclear ribonucleoprotein''), an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex that catalyzes the excision of introns from pre-mRNA. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification and takes place only in the nucleus of eukaryotes. The RNA sequence of U6 is the most highly conserved across species of all five of the snRNAs involved in the spliceosome, suggesting that the function of the U6 snRNA has remained both crucial and unchanged through evolution. It is common in vertebrate genomes to find many copies of the U6 snRNA gene or U6-derived pseudogenes. This prevalence of "back-ups" of the U6 snRNA gene in vertebrates further implies its evolutionary importance to organism viability. The U6 snRNA gene has been isolated in many org ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
U5 Spliceosomal RNA
U5 snRNA is a small nuclear RNA (snRNA) that participates in RNA splicing as a component of the spliceosome. It forms the U5 snRNP (''small nuclear ribonucleoprotein'') by associating with several proteins including Prp8 - the largest and most conserved protein in the spliceosome, Brr2 - a helicase Helicases are a class of enzymes that are vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic double helix, separating the two hybridized ... required for spliceosome activation, Snu114, and the 7 Sm proteins. U5 snRNA forms a coaxially-stacked series of helices that project into the active site of the spliceosome. Loop 1, which caps this series of helices, forms 4-5 base pairs with the 5'-exon during the two chemical reactions of splicing. This interaction appears to be especially important during step two of splicing, exon ligation. References Further reading * * ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
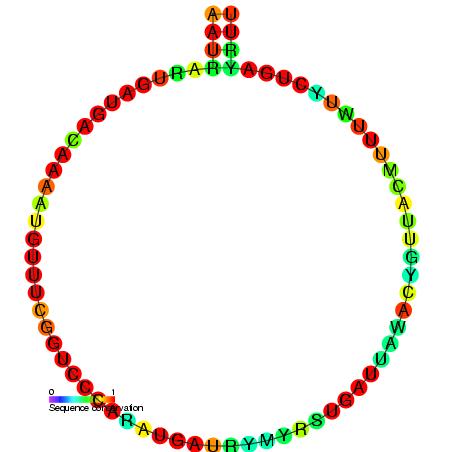
U4 Spliceosomal RNA
The U4 small nuclear Ribo-Nucleic Acid (U4 snRNA) is a non-coding RNA component of the major or U2-dependent spliceosome – a eukaryotic molecular machine involved in the splicing of pre-messenger RNA (pre-mRNA). It forms a duplex with U6 spliceosomal RNA, U6, and with each splicing round, it is displaced from the U6 snRNA (and the spliceosome) in an ATP-dependent manner, allowing U6 to re-fold and create the active site for RNA splicing, splicing catalysis. A recycling process involving protein Brr2 releases U4 from U6, while protein Prp24 re-anneals U4 and U6. The crystal structure of a 5′ stem-loop of U4 in complex with a binding protein has been solved. Biological role The U4 snRNA has been shown to exist in a number of different formats including: bound to proteins as a small nuclear Ribo-Nuclear Protein snRNP, involved with the U6 snRNA in the di-snRNP, as well as involved with both the U6 snRNA and the U5 snRNA in the tri-snRNP. The different formats have been propose ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
U2 Spliceosomal RNA
U2 spliceosomal snRNAs are a species of small nuclear RNA (snRNA) molecules found in the major spliceosomal (Sm) machinery of virtually all eukaryotic organisms. ''In vivo'', U2 snRNA along with its associated polypeptides assemble to produce the U2 small nuclear ribonucleoprotein (snRNP), an essential component of the major spliceosomal complex. The major spliceosomal-splicing pathway is occasionally referred to as U2 dependent, based on a class of Sm intron—found in mRNA primary transcripts—that are recognized exclusively by the U2 snRNP during early stages of spliceosomal assembly. In addition to U2 dependent intron recognition, U2 snRNA has been theorized to serve a catalytic role in the chemistry of pre-RNA splicing as well. Similar to ribosomal RNAs ( rRNAs), Sm snRNAs must mediate both RNA:RNA and RNA:protein contacts and hence have evolved specialized, highly conserved, primary and secondary structural elements to facilitate these types of interactions. Shortly after t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
U1 Spliceosomal RNA
U1 spliceosomal RNA is the small nuclear RNA (snRNA) component of U1 snRNP (''small nuclear ribonucleoprotein''), an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification, and takes place only in the nucleus of eukaryotes. Structure and function In humans, the U1 spliceosomal RNA is 164 bases long, forms four stem-loops, and possesses a 5'-trimethylguanosine five-prime cap. Bases 3 to 10 are a conserved sequence that base-pairs with the 5' splice site of introns during RNA splicing, and bases 126 to 133 form the Sm site, around which the Sm ring is assembled. Stem-loop I binds to the U1-70K protein, stem-loop II binds to the U1 A protein, stem-loops III and IV bind to the core RNP domain, a heteroheptameric Sm ring consisting of SmB/B', ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SnoRNA
In molecular biology, small nucleolar RNAs (snoRNAs) are a class of small RNA molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs and small nuclear RNAs. There are two main classes of snoRNA, the C/D box snoRNAs, which are associated with methylation, and the H/ACA box snoRNAs, which are associated with pseudouridylation. SnoRNAs are commonly referred to as guide RNAs but should not be confused with the guide RNAs that direct RNA editing in trypanosomes or the guide RNAs (gRNAs) used by Cas9 for CRISPR gene editing. snoRNA guided modifications After transcription, nascent rRNA molecules (termed pre-rRNA) undergo a series of processing steps to generate the mature rRNA molecule. Prior to cleavage by exo- and endonucleases, the pre-rRNA undergoes a complex pattern of nucleoside modifications. These include methylations and pseudouridylations, guided by snoRNAs. *Methylation is the attachment or substitution of a methyl group ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Motif
In biology, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and usually assumed to be related to biological function of the macromolecule. For example, an ''N''-glycosylation site motif can be defined as ''Asn, followed by anything but Pro, followed by either Ser or Thr, followed by anything but Pro residue''. Overview When a sequence motif appears in the exon of a gene, it may encode the " structural motif" of a protein; that is a stereotypical element of the overall structure of the protein. Nevertheless, motifs need not be associated with a distinctive secondary structure. " Noncoding" sequences are not translated into proteins, and nucleic acids with such motifs need not deviate from the typical shape (e.g. the "B-form" DNA double helix). Outside of gene exons, there exist regulatory sequence motifs and motifs within the " junk", such as satellite DNA. Some of these are believed to affect the shape of nucleic acids (see for example ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Three Prime End
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide pentose-sugar-ring means that there will be a 5′ end (usually pronounced "five-prime end"), which frequently contains a phosphate group attached to the 5′ carbon of the ribose ring, and a 3′ end (usually pronounced "three-prime end"), which typically is unmodified from the ribose -OH substituent. In a DNA double helix, the strands run in opposite directions to permit base pairing between them, which is essential for replication or transcription of the encoded information. Nucleic acids can only be synthesized in vivo in the 5′-to-3′ direction, as the polymerases that assemble various types of new strands generally rely on the energy produced by breaking nucleoside triphosphate bonds to attach new nucleoside monophosphates to the 3′- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |