|
Avian Metaavulavirus 2
''Avian metaavulavirus 2'', formerly ''Avian paramyxovirus 2'', is a species of virus belonging to the family ''Paramyxoviridae'' and genus ''Metaavulavirus''. The virus is a negative strand RNA virus containing a monopartite genome. ''Avian metaavulavirus 2'' is one of nine species belonging to the genus ''Metaavulavirus''. The most common serotype of ''Avulavirinae'' is serotype 1, the cause of Newcastle disease (ND). ''Avian metaavulavirus 2'' has been known to cause disease, specifically mild respiratory infections in domestic poultry, including domestic turkey, turkeys and chickens, and has many economic effects on egg production and Poultry farming, poultry industries. The virus was first isolated from a strain in Yucaipa, California in 1956. Since then, other isolates of the virus have been isolated worldwide. Viral classification ''Avian metaavulavirus 2'' is a negative sense RNA virus with a monopartite genome. It belongs to Baltimore classification#Group V: Single- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
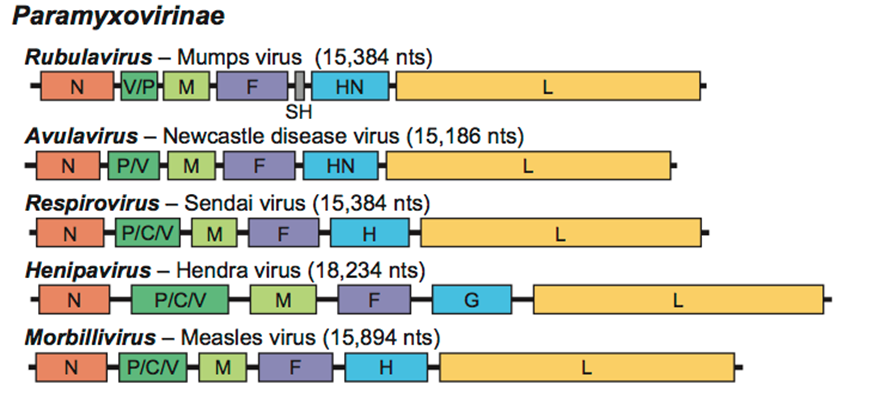
Paramyxoviridae
''Paramyxoviridae'' (from Ancient Greek, Greek ''para-'' “by the side of” and ''myxa'' “mucus”) is a family of negative-strand RNA viruses in the order ''Mononegavirales''. Vertebrates serve as natural hosts. Diseases associated with this family include measles, mumps, and respiratory tract infections. The family has four subfamilies, 17 genera, and 78 species, three genera of which are unassigned to a subfamily. Structure Virions are enveloped and can be spherical or pleomorphic and capable of producing filamentous virions. The diameter is around 150 nm. Genomes are linear, around 15kb in length. Fusion proteins and attachment proteins appear as spikes on the virion surface. Matrix proteins inside the envelope stabilise virus structure. The nucleocapsid core is composed of the genomic RNA, nucleocapsid proteins, phosphoproteins and polymerase proteins. Genome The genome is nonsegmented, negative-sense RNA, 15–19 kilobases in length, and contains six to 10 gene ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-stranded RNA Viruses - Negative-sense
When referring to DNA transcription, the coding strand (or informational strand) is the DNA strand whose base sequence is identical to the base sequence of the RNA transcript produced (although with thymine replaced by uracil). It is this strand which contains codons, while the non-coding strand contains anticodons. During transcription, RNA Pol II binds to the non-coding template strand, reads the anti-codons, and transcribes their sequence to synthesize an RNA transcript with complementary bases. By convention, the coding strand is the strand used when displaying a DNA sequence. It is presented in the 5' to 3' direction. Wherever a gene exists on a DNA molecule, one strand is the coding strand (or sense strand), and the other is the noncoding strand (also called the antisense strand, anticoding strand, template strand or transcribed strand). Strands in transcription bubble During transcription, RNA polymerase In molecular biology, RNA polymerase (abbreviated R ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polyglutamine Tract
A polyglutamine tract or polyQ tract is a portion of a protein consisting of a sequence of several glutamine units. A tract typically consists of about 10 to a few hundred such units. A multitude of genes, in various eukaryotic species (including humans), contain a number of repetitions of the nucleotide triplet C A G or C A A. When the gene is translated into a protein, each of these triplets gives rise to a glutamine unit, resulting in a polyglutamine tract. Different alleles of such a gene often have different numbers of triplets since the highly repetitive sequence is prone to contraction and expansion. Several inheritable neurodegenerative disorders, the polyglutamine diseases, occur if a mutation causes a polyglutamine tract in a specific gene to become too long. Important examples of polyglutamine diseases are spinocerebellar ataxia and Huntington's disease. Trinucleotide repeat expansion occurring in a parental germline cell can lead to children that are more affected or ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Subgenomic MRNA
Subgenomic mRNAs are essentially smaller sections of the original transcribed template strand. 3' to 5' DNA or RNA During transcription, the original template strand is usually read from the 3' to the 5' end from beginning to end. Subgenomic mRNAs are created when transcription begins at the 3' end of the template strand (or 5' of the to-be-newly synthesized template) and begins to copy towards the 5' end of the template strand before "jumping" to the end of the template and copying the last nucleotides of the 5' end of the template, (finishing the 3' tail for the newly created strand). As a result, the translated strand will have a similar 5' end to varying degrees with the original template (depending on which part of the template the transcription jumped over) and a similar 3' end to the template. 5' to 3' (positive sense) viral RNA Positive-sense (5' to 3') viral RNA which may be directly translated into the desired viral proteins, undergoes a similar process as described ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (genetics)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Promoters control gene expression in bacteria and eukaryotes. RNA polymerase must attach to DNA near a gene for transcription to occur. Promoter DNA sequences provide an enzyme binding site. The -10 sequence is TATAAT. -35 sequences are conserved on average, but not in most promoters. Artificial promoters with conserved -10 and -35 elements transcribe more slowly. All D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cis-acting Elements
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology. CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes (pleiotropy). The Latin prefix ''cis'' means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed. CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as ''cis'' because they ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetic Tree Based On The N Protein Sequences Of Selected Paramyxoviruses
In biology, phylogenetics (; from Greek φυλή/ φῦλον [] "tribe, clan, race", and wikt:γενετικός, γενετικός [] "origin, source, birth") is the study of the evolutionary history and relationships among or within groups of organisms. These relationships are determined by Computational phylogenetics, phylogenetic inference methods that focus on observed heritable traits, such as DNA sequences, protein amino acid sequences, or morphology. The result of such an analysis is a phylogenetic tree—a diagram containing a hypothesis of relationships that reflects the evolutionary history of a group of organisms. The tips of a phylogenetic tree can be living taxa or fossils, and represent the "end" or the present time in an evolutionary lineage. A phylogenetic diagram can be rooted or unrooted. A rooted tree diagram indicates the hypothetical common ancestor of the tree. An unrooted tree diagram (a network) makes no assumption about the ancestral line, and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glycoprotein
Glycoproteins are proteins which contain oligosaccharide chains covalently attached to amino acid side-chains. The carbohydrate is attached to the protein in a cotranslational or posttranslational modification. This process is known as glycosylation. Secreted extracellular proteins are often glycosylated. In proteins that have segments extending extracellularly, the extracellular segments are also often glycosylated. Glycoproteins are also often important integral membrane proteins, where they play a role in cell–cell interactions. It is important to distinguish endoplasmic reticulum-based glycosylation of the secretory system from reversible cytosolic-nuclear glycosylation. Glycoproteins of the cytosol and nucleus can be modified through the reversible addition of a single GlcNAc residue that is considered reciprocal to phosphorylation and the functions of these are likely to be an additional regulatory mechanism that controls phosphorylation-based signalling. In contrast, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription (biology)
Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1–3% of total RNA samples. Less than 2% of the human genome can be transcribed into mRNA ( Human genome#Coding vs. noncoding DNA), while at least 80% of mammalian genomic DNA can be actively transcribed (in one or more types of cells), with the majority of this 80% considered to be ncRNA. Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript. Transcription proceeds in the following general steps: # RNA polymerase, together with one or more general transcription factors, binds to promoter ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intergenic Region
An intergenic region is a stretch of DNA sequences located between genes. Intergenic regions may contain functional elements and junk DNA. ''Inter''genic regions should not be confused with ''intra''genic regions (or introns), which are non-coding regions that are found ''within'' genes, especially within the genes of eukaryotic organisms. Properties and functions Intergenic regions may contain a number of functional DNA sequences such as promoters and regulatory elements, enhancers, spacers, and (in eukaryotes) centromeres. They may also contain origins of replication, scaffold attachment regions, and transposons and viruses. Non-functional DNA elements such as pseudogenes and repetitive DNA, both of which are types of junk DNA, can also be found in intergenic regions—although they may also be located within genes in introns. As all scientific knowledge is ultimately tentative—and in principle subject to revision given better evidence—it is possible s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electron Microscope
An electron microscope is a microscope that uses a beam of accelerated electrons as a source of illumination. As the wavelength of an electron can be up to 100,000 times shorter than that of visible light photons, electron microscopes have a higher resolving power than light microscopes and can reveal the structure of smaller objects. A scanning transmission electron microscope has achieved better than 50 pm resolution in annular dark-field imaging mode and magnifications of up to about 10,000,000× whereas most light microscopes are limited by diffraction to about 200 nm resolution and useful magnifications below 2000×. Electron microscopes use shaped magnetic fields to form electron optical lens systems that are analogous to the glass lenses of an optical light microscope. Electron microscopes are used to investigate the ultrastructure of a wide range of biological and inorganic specimens including microorganisms, cells, large molecules, biopsy samples, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein. mRNA is created during the process of transcription, where an enzyme (RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and, utilising amino acids carried by transfer RNA (tRNA), the ribosome creates the protein. This process is known as translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genetic information in a biological system. As in DNA, genetic inf ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |