|
Amplified Fragment Length Polymorphism
AFLP-PCR or just AFLP is a PCR-based tool used in genetics research, DNA fingerprinting, and in the practice of genetic engineering. Developed in the early 1990s by KeyGene, AFLP uses restriction enzymes to digest genomic DNA, followed by ligation of adaptors to the sticky ends of the restriction fragments. A subset of the restriction fragments is then selected to be amplified. This selection is achieved by using primers complementary to the adaptor sequence, the restriction site sequence and a few nucleotides inside the restriction site fragments (as described in detail below). The amplified fragments are separated and visualized on denaturing on agarose gel electrophoresis , either through autoradiography or fluorescence methodologies, or via automated capillary sequencing instruments. Although AFLP should not be used as an acronym, it is commonly referred to as "Amplified fragment length polymorphism". However, the resulting data are not scored as length polymorphisms, b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
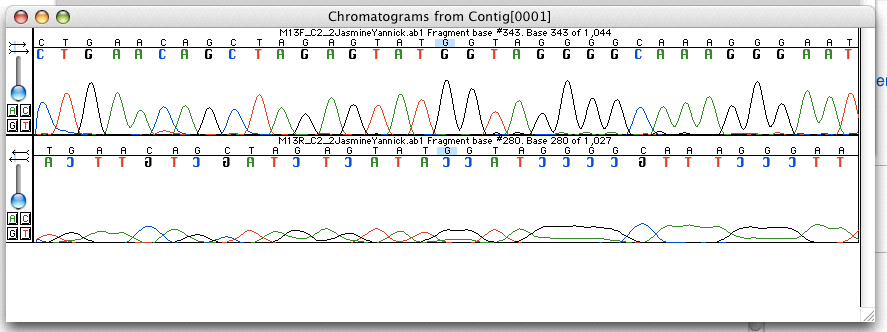
Electropherogram Trace
An electropherogram, or electrophoregram, can also be referred to as an EPG or e-gram. It is a record or chart produced when electrophoresis is used in an analytical technique, primarily in the fields of forensic biology, molecular biology and biochemistry. The method utilizes data points that correspond with a specific time and fluorescence intensity at various wavelengths of light to represent a DNA profile. In the field of genetics, an electropherogram is a plot of DNA fragment sizes, typically used for genotyping such as DNA sequencing. The data is plotted with time, shown via base pairs (bps), on the x-axis and fluorescence intensity on the y-axis. Such plots are often achieved using an instrument such as an automated DNA sequencer paired with capillary electrophoresis (CE). Such electropherograms may be used to determine DNA sequence genotypes, or genotypes that are based on the length of specific DNA fragments or number of short tandem repeats (STR) at a specific locus ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Autoradiography
An autoradiograph is an image on an X-ray film or nuclear emulsion produced by the pattern of decay emissions (e.g., beta particles or gamma rays) from a distribution of a radioactive substance. Alternatively, the autoradiograph is also available as a digital image (digital autoradiography), due to the recent development of scintillation gas detectors or rare earth phosphorimaging systems. The film or emulsion is apposed to the labeled tissue section to obtain the autoradiograph (also called an autoradiogram). The ''auto-'' prefix indicates that the radioactive substance is within the sample, as distinguished from the case of historadiography or microradiography, in which the sample is marked using an external source. Some autoradiographs can be examined microscopically for localization of silver grains (such as on the interiors or exteriors of cells or organelles) in which the process is termed micro-autoradiography. For example, micro-autoradiography was used to examine wheth ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microsatellite (genetics)
A microsatellite is a tract of repetitive DNA in which certain DNA motifs (ranging in length from one to six or more base pairs) are repeated, typically 5–50 times. Microsatellites occur at thousands of locations within an organism's genome. They have a higher mutation rate than other areas of DNA leading to high genetic diversity. Microsatellites are often referred to as short tandem repeats (STRs) by forensic geneticists and in genetic genealogy, or as simple sequence repeats (SSRs) by plant geneticists. Microsatellites and their longer cousins, the minisatellites, together are classified as VNTR (variable number of tandem repeats) DNA. The name "satellite" DNA refers to the early observation that centrifugation of genomic DNA in a test tube separates a prominent layer of bulk DNA from accompanying "satellite" layers of repetitive DNA. They are widely used for DNA profiling in cancer diagnosis, in kinship analysis (especially paternity testing) and in forensic identi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Random Amplification Of Polymorphic DNA
Random amplification of polymorphic DNA (RAPD), pronounced "rapid", is a type of polymerase chain reaction (PCR), but the segments of DNA that are amplified are random. The scientist performing RAPD creates several arbitrary, short primers (10- 12 nucleotides), then proceeds with the PCR using a large template of genomic DNA, hoping that fragments will amplify. By resolving the resulting patterns, a semi-unique profile can be gleaned from an RAPD reaction. No knowledge of the DNA sequence of the targeted genome is required, as the primers will bind somewhere in the sequence, but it is not certain exactly where. This makes the method popular for comparing the DNA of biological systems that have not had the attention of the scientific community, or in a system in which relatively few DNA sequences are compared (it is not suitable for forming a cDNA databank). Because it relies on a large, intact DNA template sequence, it has some limitations in the use of degraded DNA samples. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Quantitative Trait Locus
A quantitative trait locus (QTL) is a locus (section of DNA) that correlates with variation of a quantitative trait in the phenotype of a population of organisms. QTLs are mapped by identifying which molecular markers (such as SNPs or AFLPs) correlate with an observed trait. This is often an early step in identifying the actual genes that cause the trait variation. Definition A quantitative trait locus (QTL) is a region of DNA which is associated with a particular phenotypic trait, which varies in degree and which can be attributed to polygenic effects, i.e., the product of two or more genes, and their environment. . These QTLs are often found on different chromosomes. The number of QTLs which explain variation in the phenotypic trait indicates the genetic architecture of a trait. It may indicate that plant height is controlled by many genes of small effect, or by a few genes of large effect. Typically, QTLs underlie continuous traits (those traits which vary continuou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
AFLP Clustering Analysis , a life-threatening liver condition that may occur during pregnancy
{{disambig ...
AFLP may refer to: *Amplified fragment length polymorphism, a highly sensitive tool used in molecular biology to detect DNA polymorphisms *Acute fatty liver of pregnancy Acute fatty liver of pregnancy is a rare life-threatening complication of pregnancy that occurs in the third trimester or the immediate period after delivery. It is thought to be caused by a disordered metabolism of fatty acids by mitochondria in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amplicon
In molecular biology, an amplicon is a piece of DNA or RNA that is the source and/or product of amplification or replication events. It can be formed artificially, using various methods including polymerase chain reactions (PCR) or ligase chain reactions (LCR), or naturally through gene duplication. In this context, ''amplification'' refers to the production of one or more copies of a genetic fragment or target sequence, specifically the amplicon. As it refers to the product of an amplification reaction, ''amplicon'' is used interchangeably with common laboratory terms, such as "PCR product." Artificial amplification is used in research, forensics, and medicine for purposes that include detection and quantification of infectious agents, identification of human remains, and extracting genotypes from human hair. Natural gene duplication plays a major role in evolution. It is also implicated in several forms of human cancer including primary mediastinal B cell lymphoma and Hodgkin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gel Electrophoresis Of Nucleic Acids
Nucleic acid electrophoresis is an analytical technique used to separate DNA or RNA fragments by size and reactivity. Nucleic acid molecules which are to be analyzed are set upon a viscous medium, the gel, where an electric field induces the nucleic acids (which are negatively charged due to their sugar- phosphate backbone) to migrate toward the anode (which is positively charged because this is an electrolytic rather than galvanic cell). The separation of these fragments is accomplished by exploiting the mobilities with which different sized molecules are able to pass through the gel. Longer molecules migrate more slowly because they experience more resistance within the gel. Because the size of the molecule affects its mobility, smaller fragments end up nearer to the anode than longer ones in a given period. After some time, the voltage is removed and the fragmentation gradient is analyzed. For larger separations between similar sized fragments, either the voltage or run time ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone (i.e. each strand) of the DNA double helix. These enzymes are found in bacteria and archaea and provide a defense mechanism against invading viruses. Inside a prokaryote, the restriction enzymes selectively cut up ''foreign'' DNA in a process called ''restriction digestion''; meanwhile, host DNA is protected by a modification enzyme (a methyltransferase) that modi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymorphism (biology)
In biology, polymorphism is the occurrence of two or more clearly different morphs or forms, also referred to as alternative ''phenotypes'', in the population of a species. To be classified as such, morphs must occupy the same habitat at the same time and belong to a panmictic population (one with random mating). Ford E.B. 1965. ''Genetic polymorphism''. Faber & Faber, London. Put simply, polymorphism is when there are two or more possibilities of a trait on a gene. For example, there is more than one possible trait in terms of a jaguar's skin colouring; they can be light morph or dark morph. Due to having more than one possible variation for this gene, it is termed 'polymorphism'. However, if the jaguar has only one possible trait for that gene, it would be termed "monomorphic". For example, if there was only one possible skin colour that a jaguar could have, it would be termed monomorphic. The term polyphenism can be used to clarify that the different forms arise from the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fluorescence
Fluorescence is the emission of light by a substance that has absorbed light or other electromagnetic radiation. It is a form of luminescence. In most cases, the emitted light has a longer wavelength, and therefore a lower photon energy, than the absorbed radiation. A perceptible example of fluorescence occurs when the absorbed radiation is in the ultraviolet region of the electromagnetic spectrum (invisible to the human eye), while the emitted light is in the visible region; this gives the fluorescent substance a distinct color that can only be seen when the substance has been exposed to UV light. Fluorescent materials cease to glow nearly immediately when the radiation source stops, unlike phosphorescent materials, which continue to emit light for some time after. Fluorescence has many practical applications, including mineralogy, gemology, medicine, chemical sensors ( fluorescence spectroscopy), fluorescent labelling, dyes, biological detectors, cosmic-ray detec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |