|
Algae DNA Barcoding
DNA barcoding of algae is commonly used for species identification and phylogenetic studies. Algae form a phylogenetically heterogeneous group, meaning that the application of a single universal barcode/ marker for species delimitation is unfeasible, thus different markers/barcodes are applied for this aim in different algal groups. Diatoms Diatom DNA barcoding is a method for taxonomical identification of diatoms even to species level. It is conducted using DNA or RNA followed by amplification and sequencing of specific, conserved regions in the diatom genome followed by taxonomic assignment. One of the main challenges of identifying diatoms is that it is often collected as a mixture of diatoms from several species. DNA metabarcoding is the process of identifying the individual species from a mixed sample of environmental DNA (also called eDNA) which is DNA extracted straight from the environment such as in soil or water samples. A newly applied method is diatom DNA m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Barcoding
DNA barcoding is a method of species identification using a short section of DNA from a specific gene or genes. The premise of DNA barcoding is that by comparison with a reference library of such DNA sections (also called "sequences"), an individual sequence can be used to uniquely identify an organism to species, just as a supermarket scanner uses the familiar black stripes of the UPC barcode to identify an item in its stock against its reference database. These "barcodes" are sometimes used in an effort to identify unknown species or parts of an organism, simply to catalog as many taxa as possible, or to compare with traditional taxonomy in an effort to determine species boundaries. Different gene regions are used to identify the different organismal groups using barcoding. The most commonly used barcode region for animals and some protists is a portion of the cytochrome ''c'' oxidase I (COI or COX1) gene, found in mitochondrial DNA. Other genes suitable for DNA barcoding ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Morphology (biology)
Morphology is a branch of biology dealing with the study of the form and structure of organisms and their specific structural features. This includes aspects of the outward appearance (shape, structure, colour, pattern, size), i.e. external morphology (or eidonomy), as well as the form and structure of the internal parts like bones and organs, i.e. internal morphology (or anatomy). This is in contrast to physiology, which deals primarily with function. Morphology is a branch of life science dealing with the study of gross structure of an organism or taxon and its component parts. History The etymology of the word "morphology" is from the Ancient Greek (), meaning "form", and (), meaning "word, study, research". While the concept of form in biology, opposed to function, dates back to Aristotle (see Aristotle's biology), the field of morphology was developed by Johann Wolfgang von Goethe (1790) and independently by the German anatomist and physiologist Karl Friedrich Burdach ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
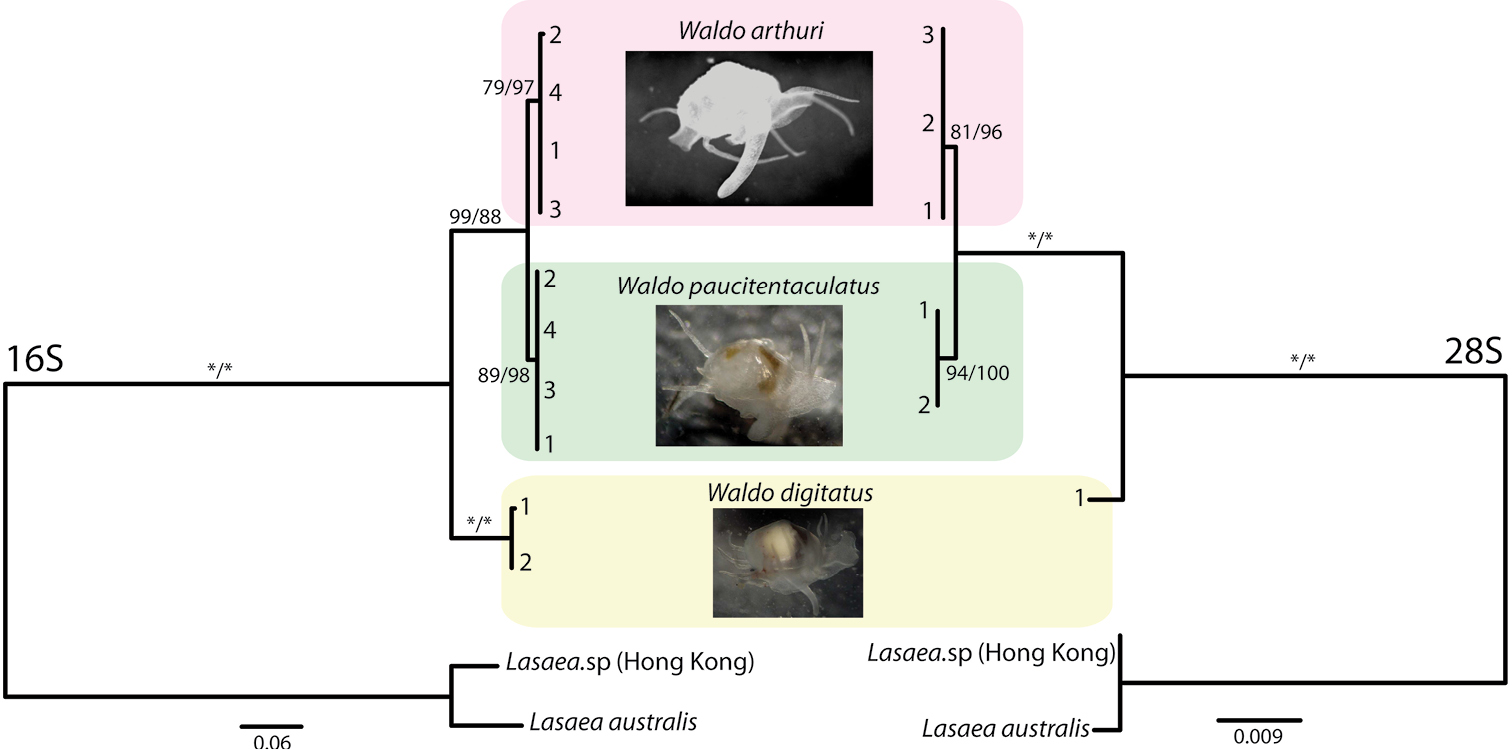
Jan Pawlowski Et Al Abstract 2018
Jan, JaN or JAN may refer to: Acronyms * Jackson, Mississippi (Amtrak station), US, Amtrak station code JAN * Jackson-Evers International Airport, Mississippi, US, IATA code * Jabhat al-Nusra (JaN), a Syrian militant group * Japanese Article Number, a barcode standard compatible with EAN * Japanese Accepted Name, a Japanese nonproprietary drug name * Job Accommodation Network, US, for people with disabilities * ''Joint Army-Navy'', US standards for electronic color codes, etc. * ''Journal of Advanced Nursing'' Personal name * Jan (name), male variant of ''John'', female shortened form of ''Janet'' and ''Janice'' * Jan (Persian name), Persian word meaning 'life', 'soul', 'dear'; also used as a name * Ran (surname), romanized from Mandarin as Jan in Wade–Giles * Ján, Slovak name Other uses * January, as an abbreviation for the first month of the year in the Gregorian calendar * Jan (cards), a term in some card games when a player loses without taking any tricks or scoring a m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosomal RNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form SSU rRNA, small and LSU rRNA, large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and Translation (biology), translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins by mass. Structure Although the primary structure of rRNA sequences can vary across organisms, Base pair, base-pairing within these sequences commonly forms stem-loop configurations. The length and position of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Taxonomic Resolution
DNA barcoding is a method of species identification using a short section of DNA from a specific gene or genes. The premise of DNA barcoding is that by comparison with a reference library of such DNA sections (also called "DNA sequence, sequences"), an individual sequence can be used to uniquely identify an organism to species, just as a supermarket scanner uses the familiar black stripes of the Universal Product Code, UPC barcode to identify an item in its stock against its reference database. These "barcodes" are sometimes used in an effort to identify unknown species or parts of an organism, simply to catalog as many Taxon, taxa as possible, or to compare with traditional taxonomy in an effort to determine species boundaries. Different gene regions are used to identify the different organismal groups using barcoding. The most commonly used barcode region for animals and some protists is a portion of the Cytochrome c oxidase subunit I, cytochrome ''c'' oxidase I (COI or Cytochr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Invertebrate
Invertebrates are a paraphyletic group of animals that neither possess nor develop a vertebral column (commonly known as a ''backbone'' or ''spine''), derived from the notochord. This is a grouping including all animals apart from the chordate subphylum Vertebrata. Familiar examples of invertebrates include arthropods, mollusks, annelids, echinoderms and cnidarians. The majority of animal species are invertebrates; one estimate puts the figure at 97%. Many invertebrate taxa have a greater number and variety of species than the entire subphylum of Vertebrata. Invertebrates vary widely in size, from 50 μm (0.002 in) rotifers to the 9–10 m (30–33 ft) colossal squid. Some so-called invertebrates, such as the Tunicata and Cephalochordata, are more closely related to vertebrates than to other invertebrates. This makes the invertebrates paraphyletic, so the term has little meaning in taxonomy. Etymology The word "invertebrate" comes from the Latin word ''vertebra'', whi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
28S Ribosomal RNA
28S ribosomal RNA is the structural ribosomal RNA (rRNA) for the large subunit (LSU) of eukaryotic cytoplasmic ribosomes, and thus one of the basic components of all eukaryotic cells. It has a size of 25S in plants and 28S in mammals, hence the alias of 25S–28S rRNA. Combined with 5.8S rRNA to the 5' side, it is the eukaryotic nuclear homologue of the prokaryotic 23S and mitochondrial 16S ribosomal RNAs. Use in phylogeny The genes coding for 28S rRNA are referred to as 28S rDNA. The comparison of the sequences from these genes are sometimes used in molecular analysis to construct phylogenetic trees, for example in protists, fungi, insects, arachnids, tardigrades, and vertebrates. Structure The 28S rRNA is typically 4000–5000 nt long. Some eukaryotes cleave 28S rRNA into two parts before assembling both into the ribosome, a phenomenon termed the "hidden break". Databases Several databases provide alignments and annotations of LSU rRNA sequences for comparati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
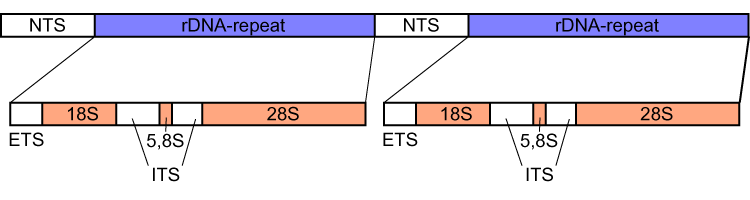
Internal Transcribed Spacer
Internal transcribed spacer (ITS) is the spacer DNA situated between the small-subunit ribosomal RNA (rRNA) and large-subunit rRNA genes in the chromosome or the corresponding transcribed region in the polycistronic rRNA precursor transcript. ITS across life domains In bacteria and archaea, there is a single ITS, located between the 16S and 23S rRNA genes. Conversely, there are two ITSs in eukaryotes: ITS1 is located between 18S and 5.8S rRNA genes, while ITS2 is between 5.8S and 28S (in opisthokonts, or 25S in plants) rRNA genes. ITS1 corresponds to the ITS in bacteria and archaea, while ITS2 originated as an insertion that interrupted the ancestral 23S rRNA gene. Organization In bacteria and archaea, the ITS occurs in one to several copies, as do the flanking 16S and 23S genes. When there are multiple copies, these do not occur adjacent to one another. Rather, they occur in discrete locations in the circular chromosome. It is not uncommon in bacteria to carry tRN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytochrome C Oxidase Subunit I
Cytochrome c oxidase I (COX1) also known as mitochondrially encoded cytochrome c oxidase I (MT-CO1) is a protein that in humans is encoded by the ''MT-CO1'' gene. In other eukaryotes, the gene is called ''COX1'', ''CO1'', or ''COI''. Cytochrome c oxidase I is the main subunit of the cytochrome c oxidase complex. Mutations in MT-CO1 have been associated with Leber's hereditary optic neuropathy (LHON), acquired idiopathic sideroblastic anemia, Complex IV deficiency, colorectal cancer, sensorineural deafness, and recurrent myoglobinuria. Structure One of 37 mitochondrial genes, the ''MT-CO1'' gene is located from nucleotide pairs 5904 to 7444 on the guanine-rich heavy (H) section of mtDNA. The gene product is a 57 kDa protein composed of 513 amino acids. Function Cytochrome c oxidase subunit I (CO1 or MT-CO1) is one of three mitochondrial DNA (mtDNA) encoded subunits (MT-CO1, MT-CO2, MT-CO3) of respiratory complex IV. Complex IV is the third and final enzyme of the electron ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RuBisCO
Ribulose-1,5-bisphosphate carboxylase-oxygenase, commonly known by the abbreviations RuBisCo, rubisco, RuBPCase, or RuBPco, is an enzyme () involved in the first major step of carbon fixation, a process by which atmospheric carbon dioxide is converted by plants and other photosynthesis, photosynthetic organisms to fuel, energy-rich molecules such as glucose. In chemical terms, it catalysis, catalyzes the carboxylation of ribulose-1,5-bisphosphate (also known as RuBP). It is probably the most abundant enzyme on Earth. Alternative carbon fixation pathways RuBisCO is important biology, biologically because it catalyzes the primary chemical reaction by which Total inorganic carbon, inorganic carbon enters the biosphere. While many autotrophic bacteria and archaea fix carbon via the reductive acetyl CoA Pathway, reductive acetyl CoA pathway, the 3-hydroxypropionate cycle, or the reverse Krebs cycle, these pathways are relatively small contributors to global carbon fixation compared t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
18S Ribosomal RNA
18S ribosomal RNA (abbreviated 18S rRNA) is a part of the ribosomal RNA. The S in 18S represents Svedberg units. 18S rRNA is an SSU rRNA, a component of the eukaryotic ribosomal small subunit (40S). 18S rRNA is the structural RNA for the small component of eukaryotic cytoplasmic ribosomes, and thus one of the basic components of all eukaryotic cells. 18S rRNA is the eukaryotic cytosolic homologue of 16S ribosomal RNA in prokaryotes and plastids. 18S rRNA is also a homologue of 12S ribosomal RNA in mitochondria. The genes coding for 18S rRNA are referred to as 18S rRNA genes. Sequence data from these genes is widely used in molecular analysis to reconstruct the evolutionary history of organisms, especially in vertebrates, as its slow evolutionary rate makes it suitable to reconstruct ancient divergences. Uses in phylogeny The small subunit (SSU) 18S rRNA gene is one of the most frequently used genes in phylogenetic studies and an important marker for random target polymerase ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |