|
80's (radio)
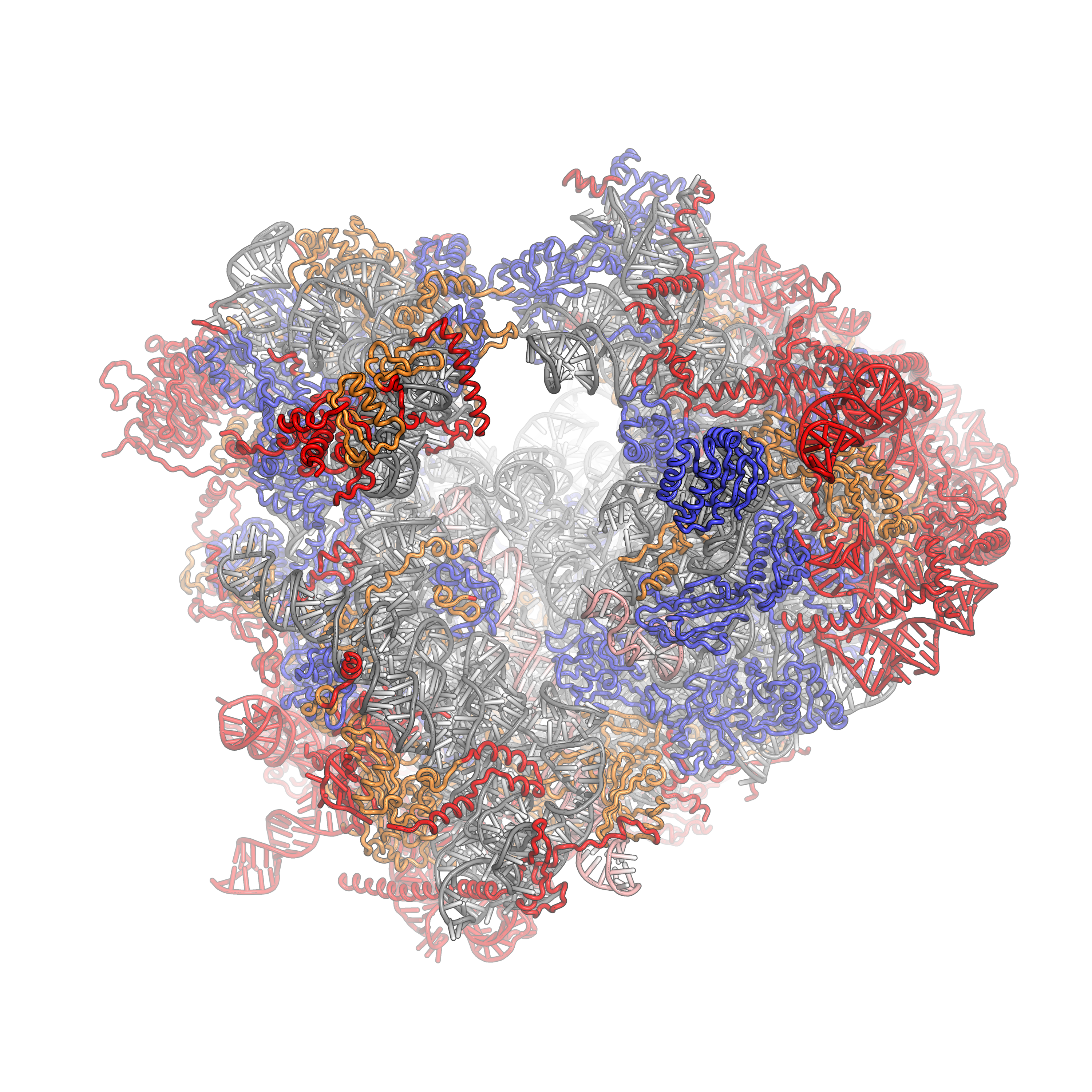
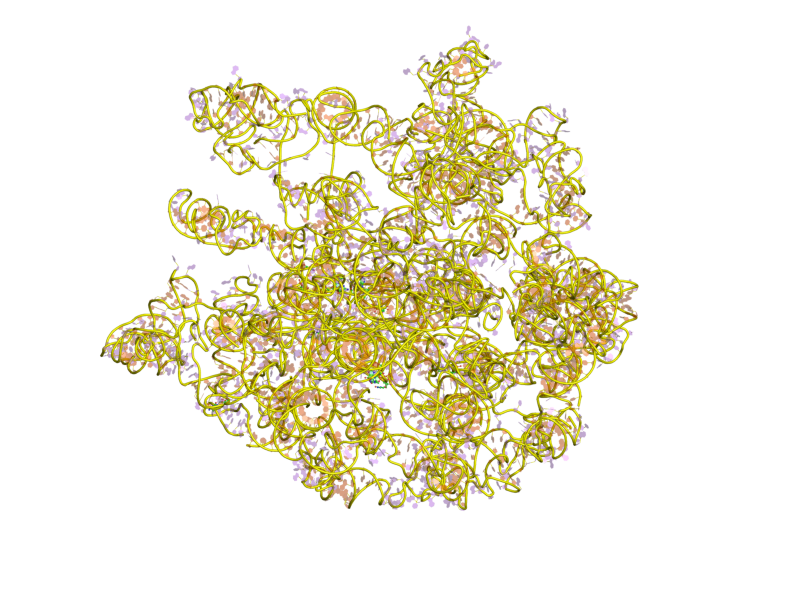
Ribosomes are a large and complex molecular machine that catalyzes the synthesis of proteins, referred to as translation. The ribosome selects aminoacylated transfer RNAs (tRNAs) based on the sequence of a protein-encoding messenger RNA (mRNA) and covalently links the amino acids into a polypeptide chain. Ribosomes from all organisms share a highly conserved catalytic center. However, the ribosomes of eukaryotes (animals, plants, fungi, and large number unicellular organisms all with a nucleus) are much larger than prokaryotic (bacterial and archaeal) ribosomes and subject to more complex regulation and biogenesis pathways. Eukaryotic ribosomes are also known as 80S ribosomes, referring to their sedimentation coefficients in Svedberg units, because they sediment faster than the prokaryotic (70S) ribosomes. Eukaryotic ribosomes have two unequal subunits, designated small subunit (40S) and large subunit (60S) according to their sedimentation coefficients. Both subunits contain doze ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
80S 2XZM 4A17 4A19
8 (eight) is the natural number following 7 and preceding 9. In mathematics 8 is: * a composite number, its proper divisors being , , and . It is twice 4 or four times 2. * a power of two, being 2 (two cubed), and is the first number of the form , being an integer greater than 1. * the first number which is neither prime nor semiprime. * the base of the octal number system, which is mostly used with computers. In octal, one digit represents three bits. In modern computers, a byte is a grouping of eight bits, also called an octet. * a Fibonacci number, being plus . The next Fibonacci number is . 8 is the only positive Fibonacci number, aside from 1, that is a perfect cube. * the only nonzero perfect power that is one less than another perfect power, by Mihăilescu's Theorem. * the order of the smallest non-abelian group all of whose subgroups are normal. * the dimension of the octonions and is the highest possible dimension of a normed division algebra. * the first number ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryotic Large Ribosomal Subunit (60S)
Ribosomal particles are denoted according to their sedimentation coefficients in Svedberg units. The 60S subunit is the large subunit of eukaryotic 80S ribosomes. It is structurally and functionally related to the 50S subunit of 70S prokaryotic ribosomes. However, the 60S subunit is much larger than the prokaryotic 50S subunit and contains many additional protein segments, as well as ribosomal RNA expansion segments. Overall structure Characteristic features of the large subunit, shown below in the "Crown View", include the central protuberance (CP) and the two stalks, which are named according to their bacterial protein components (L1 stalk on the left as seen from the subunit interface and L7/L12 on the right). There are three binding sites for tRNA, the A-site, P-site and E-site (see article on protein translation for details). The core of the 60S subunit is formed by the 28S ribosomal RNA (abbreviated 28S rRNA), which is homologous to the prokaryotic 23S rRNA, which also co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electron Microscopy
An electron microscope is a microscope that uses a beam of accelerated electrons as a source of illumination. As the wavelength of an electron can be up to 100,000 times shorter than that of visible light photons, electron microscopes have a higher resolving power than light microscopes and can reveal the structure of smaller objects. A scanning transmission electron microscope has achieved better than 50 pm resolution in annular dark-field imaging mode and magnifications of up to about 10,000,000× whereas most light microscopes are limited by diffraction to about 200 nm resolution and useful magnifications below 2000×. Electron microscopes use shaped magnetic fields to form electron optical lens systems that are analogous to the glass lenses of an optical light microscope. Electron microscopes are used to investigate the ultrastructure of a wide range of biological and inorganic specimens including microorganisms, cells, large molecules, biopsy samples, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleotide
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver. Nucleotides are composed of three subunit molecules: a nucleobase, a five-carbon sugar (ribose or deoxyribose), and a phosphate group consisting of one to three phosphates. The four nucleobases in DNA are guanine, adenine, cytosine and thymine; in RNA, uracil is used in place of thymine. Nucleotides also play a central role in metabolism at a fundamental, cellular level. They provide chemical energy—in the form of the nucleoside triphosphates, adenosine triphosphate (ATP), guanosine triphosphate (GTP), cytidine triphosphate (CTP) and uridine triphosphate (UTP)—throughout the cell for the many cellular func ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Angstrom
The angstromEntry "angstrom" in the Oxford online dictionary. Retrieved on 2019-03-02 from https://en.oxforddictionaries.com/definition/angstrom.Entry "angstrom" in the Merriam-Webster online dictionary. Retrieved on 2019-03-02 from https://www.merriam-webster.com/dictionary/angstrom. (, ; , ) or ångström is a metric unit of length equal to m; that is, one ten-billionth ( US) of a metre, a hundred-millionth of a centimetre,Entry "angstrom" in the Oxford English Dictionary, 2nd edition (1986). Retrieved on 2021-11-22 from https://www.oed.com/oed2/00008552. 0.1 nanometre, or 100 picometres. Its symbol is Å, a letter of the Swedish alphabet. The unit is named after the Swedish physicist Anders Jonas Ångström (1814–1874). The angstrom is often used in the natural sciences and technology to express sizes of atoms, molecules, microscopic biological structures, and lengths of chemical bonds, arrangement of atoms in crystals,Arturas Vailionis (2015):Geometry of Crystals Lect ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Atomic Mass Unit
The dalton or unified atomic mass unit (symbols: Da or u) is a non-SI unit of mass widely used in physics and chemistry. It is defined as of the mass of an unbound neutral atom of carbon-12 in its nuclear and electronic ground state and at rest. The atomic mass constant, denoted ''m''u, is defined identically, giving . This unit is commonly used in physics and chemistry to express the mass of atomic-scale objects, such as atoms, molecules, and elementary particles, both for discrete instances and multiple types of ensemble averages. For example, an atom of helium-4 has a mass of . This is an intrinsic property of the isotope and all helium-4 atoms have the same mass. Acetylsalicylic acid (aspirin), , has an average mass of approximately . However, there are no acetylsalicylic acid molecules with this mass. The two most common masses of individual acetylsalicylic acid molecules are , having the most common isotopes, and , in which one carbon is carbon-13. The molecular mass ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Mass
The molecular mass (''m'') is the mass of a given molecule: it is measured in daltons (Da or u). Different molecules of the same compound may have different molecular masses because they contain different isotopes of an element. The related quantity relative molecular mass, as defined by IUPAC, is the ratio of the mass of a molecule to the unified atomic mass unit (also known as the dalton) and is unitless. The molecular mass and relative molecular mass are distinct from but related to the molar mass. The molar mass is defined as the mass of a given substance divided by the amount of a substance and is expressed in g/mol. That makes the molar mass an average of many particles or molecules, and the molecular mass the mass of one specific particle or molecule. The molar mass is usually the more appropriate figure when dealing with macroscopic (weigh-able) quantities of a substance. The definition of molecular weight is most authoritatively synonymous with relative molecular mass; ho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
23S Ribosomal RNA
The 23S rRNA is a 2,904 nucleotide long (in '' E. coli'') component of the large subunit ( 50S) of the bacterial/archean ribosome and makes up the peptidyl transferase center (PTC). The 23S rRNA is divided into six secondary structural domains titled I-VI, with the corresponding 5S rRNA being considered domain VII. The ribosomal peptidyl transferase activity resides in domain V of this rRNA, which is also the most common binding site for antibiotics that inhibit translation, making it a target for ribosomal engineering. A well-known member of this antibiotic class, chloramphenicol, acts by inhibiting peptide bond formation, with recent 3D-structural studies showing two different binding sites depending on the species of ribosome. Numerous mutations in domains of the 23S rRNA with Peptidyl transferase activity have resulted in antibiotic resistance. 23S rRNA genes typically have higher sequence variations, including insertions and/or deletions, compared to other rRNAs. The euka ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
16S Ribosomal RNA
16 S ribosomal RNA (or 16 S rRNA) is the RNA component of the 30S subunit of a prokaryotic ribosome (SSU rRNA). It binds to the Shine-Dalgarno sequence and provides most of the SSU structure. The genes coding for it are referred to as 16S rRNA gene and are used in reconstructing phylogenies, due to the slow rates of evolution of this region of the gene. Carl Woese and George E. Fox were two of the people who pioneered the use of 16S rRNA in phylogenetics in 1977. Multiple sequences of the 16S rRNA gene can exist within a single bacterium. Functions * Like the large (23S) ribosomal RNA, it has a structural role, acting as a scaffold defining the positions of the ribosomal proteins. * The 3-end contains the anti- Shine-Dalgarno sequence, which binds upstream to the AUG start codon on the mRNA. The 3-end of 16S RNA binds to the proteins S1 and S21 which are known to be involved in initiation of protein synthesis * Interacts with 23S, aiding in the binding of the two ribosomal s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
18S Ribosomal RNA
18S ribosomal RNA (abbreviated 18S rRNA) is a part of the ribosomal RNA. The S in 18S represents Svedberg units. 18S rRNA is an SSU rRNA, a component of the eukaryotic ribosomal small subunit (40S). 18S rRNA is the structural RNA for the small component of eukaryotic cytoplasmic ribosomes, and thus one of the basic components of all eukaryotic cells. 18S rRNA is the eukaryotic cytosolic homologue of 16S ribosomal RNA in prokaryotes and plastids. 18S rRNA is also a homologue of 12S ribosomal RNA in mitochondria. The genes coding for 18S rRNA are referred to as 18S rRNA genes. Sequence data from these genes is widely used in molecular analysis to reconstruct the evolutionary history of organisms, especially in vertebrates, as its slow evolutionary rate makes it suitable to reconstruct ancient divergences. Uses in phylogeny The small subunit (SSU) 18S rRNA gene is one of the most frequently used genes in phylogenetic studies and an important marker for random target polymerase ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
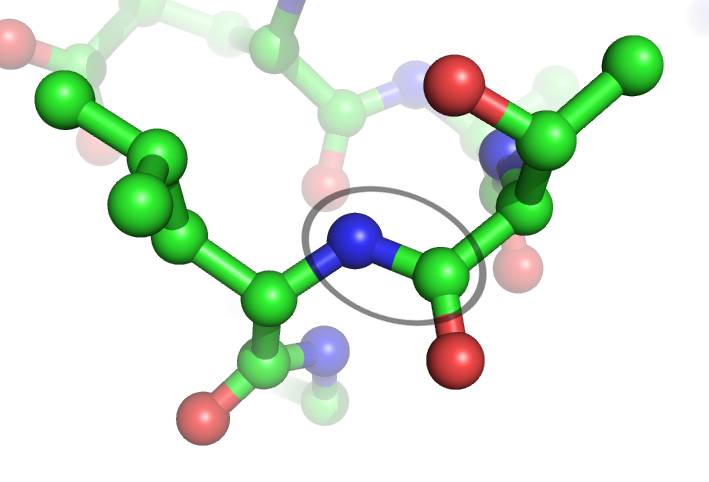
Peptide Bond
In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 (carbon number one) of one alpha-amino acid and N2 (nitrogen number two) of another, along a peptide or protein chain. It can also be called a eupeptide bond to distinguish it from an isopeptide bond, which is another type of amide bond between two amino acids. Synthesis When two amino acids form a ''dipeptide'' through a ''peptide bond'', it is a type of condensation reaction. In this kind of condensation, two amino acids approach each other, with the non-side chain (C1) carboxylic acid moiety of one coming near the non-side chain (N2) amino moiety of the other. One loses a hydrogen and oxygen from its carboxyl group (COOH) and the other loses a hydrogen from its amino group (NH2). This reaction produces a molecule of water (H2O) and two amino acids joined by a peptide bond (−CO−NH−). The two joined amino acids are called a dipeptide. The am ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Anticodon
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino acid sequence of proteins. tRNAs genes from Bacteria are typically shorter (mean = 77.6 bp) than tRNAs from Archaea (mean = 83.1 bp) and eukaryotes (mean = 84.7 bp). The mature tRNA follows an opposite pattern with tRNAs from Bacteria being usually longer (median = 77.6 nt) than tRNAs from Archaea (median = 76.8 nt), with eukaryotes exhibiting the shortest mature tRNAs (median = 74.5 nt). Transfer RNA (tRNA) does this by carrying an amino acid to the protein synthesizing machinery of a cell called the ribosome. Complementation of a 3-nucleotide codon in a messenger RNA (mRNA) by a 3-nucleotide anticodon of the tRNA results in protein synthesis based on the mRNA code. As such, tRNAs are a necessary component of translation, the biological syn ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |