|
8-hydroxy-2'-deoxyguanosine
8-Oxo-2'-deoxyguanosine (8-oxo-dG) is an oxidized derivative of deoxyguanosine. 8-Oxo-dG is one of the major products of DNA oxidation. Concentrations of 8-oxo-dG within a cell are a measurement of oxidative stress. In DNA Steady-state levels of DNA damages represent the balance between formation and repair. Swenberg et al. measured average frequencies of steady state endogenous DNA damages in mammalian cells. The most frequent oxidative DNA damage normally present in DNA is 8-oxo-dG, occurring at an average frequency of 2,400 per cell. When 8-oxo-dG is induced by a DNA damaging agent it is rapidly repaired. For example, 8-oxo-dG was increased 10-fold in the livers of mice subjected to ionizing radiation, but the excess 8-oxo-dG was rapidly removed with a half-life of 11 minutes. As reviewed by Valavanidis et al. increased levels of 8-oxo-dG in a tissue can serve as a biomarker of oxidative stress. They also noted that increased levels of 8-oxo-dG are frequently found dur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Oxidation
DNA oxidation is the process of oxidative damage of deoxyribonucleic acid. As described in detail by Burrows et al., 8-oxo-2'-deoxyguanosine (8-oxo-dG) is the most common oxidative lesion observed in duplex DNA because guanine has a lower one-electron reduction potential than the other nucleosides in DNA. The one electron reduction potentials of the nucleosides (in volts versus NHE) are guanine 1.29, adenine 1.42, cytosine 1.6 and thymine 1.7. About 1 in 40,000 guanines in the genome are present as 8-oxo-dG under normal conditions. This means that >30,000 8-oxo-dGs may exist at any given time in the genome of a human cell. Another product of DNA oxidation is 8-oxo-dA. 8-oxo-dA occurs at about 1/10 the frequency of 8-oxo-dG. The reduction potential of guanine may be reduced by as much as 50%, depending on the particular neighboring nucleosides stacked next to it within DNA. Excess DNA oxidation is linked to certain diseases and cancers, while normal levels of oxidized nucleot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenetics In Learning And Memory
While the cellular and molecular mechanisms of learning and memory have long been a central focus of neuroscience, it is only in recent years that attention has turned to the epigenetic mechanisms behind the dynamic changes in gene transcription responsible for memory formation and maintenance. Epigenetic gene regulation often involves the physical marking (chemical modification) of DNA or associated proteins to cause or allow long-lasting changes in gene activity. Epigenetic mechanisms such as DNA methylation and histone modifications (methylation, acetylation, and deacetylation) have been shown to play an important role in learning and memory. DNA Methylation DNA methylation involves the addition of a methyl group to a 5' cytosine residue. This usually occurs at cytosines that form part of a cytosine-guanine dinucleotide (CpG sites). Methylation can lead to activation or repression of gene transcription and is mediated through the activity of DNA methyltransferases (DNMTs). DNM ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CpG Site
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG islands (or CG islands). Cytosines in CpG dinucleotides can be methylated to form 5-methylcytosines. Enzymes that add a methyl group are called DNA methyltransferases. In mammals, 70% to 80% of CpG cytosines are methylated. Methylating the cytosine within a gene can change its expression, a mechanism that is part of a larger field of science studying gene regulation that is called epigenetics. Methylated cytosines often mutate to thymines. In humans, about 70% of promoters located near the transcription start site of a gene (proximal promoters) contain a CpG island. CpG characteristics Definition ''CpG'' is shorthand for ''5'—C—phosphate—G—3' '', that is, cytosine and guanine separated by only one phosphate group; phosphate ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Damage
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA crosslinkages (interstrand crosslinks or ICLs). This can eventually lead to maligna ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Demethylation
For molecular biology in mammals, DNA demethylation causes replacement of 5-methylcytosine (5mC) in a DNA sequence by cytosine (C) (see figure of 5mC and C). DNA demethylation can occur by an active process at the site of a 5mC in a DNA sequence or, in replicating cells, by preventing addition of methyl groups to DNA so that the replicated DNA will largely have cytosine in the DNA sequence (5mC will be diluted out). Methylated cytosine is frequently present in the linear DNA sequence where a cytosine is followed by a guanine in a 5' → 3' direction (a CpG site). In mammals, DNA methyltransferases (which add methyl groups to DNA bases) exhibit a strong sequence preference for cytosines at CpG sites. There appear to be more than 20 million CpG dinucleotides in the human genome (see genomic distribution). In mammals, on average, 70% to 80% of CpG cytosines are methylated, though the level of methylation varies with different tissues. Methylated cytosines often occur in groups ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Repair
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA crosslinkages (interstrand crosslinks or ICLs). This can eventually lead to malignant ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Deoxyguanosine
Deoxyguanosine is composed of the purine nucleobase guanine linked by its N9 nitrogen to the C1 carbon of deoxyribose. It is similar to guanosine, but with one hydroxyl group removed from the 2' position of the ribose sugar (making it deoxyribose). If a phosphate group is attached at the 5' position, it becomes deoxyguanosine monophosphate. Deoxyguanosine is one of the four deoxyribonucleosides that make up DNA. See also * 8-Oxo-2'-deoxyguanosine 8-Oxo-2'-deoxyguanosine (8-oxo-dG) is an oxidized derivative of deoxyguanosine. 8-Oxo-dG is one of the major products of DNA oxidation. Concentrations of 8-oxo-dG within a cell are a measurement of oxidative stress. In DNA Steady-state leve ... References Nucleosides Purines Hydroxymethyl compounds {{Biochemistry-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methyl Group
In organic chemistry, a methyl group is an alkyl derived from methane, containing one carbon atom bonded to three hydrogen atoms, having chemical formula . In formulas, the group is often abbreviated as Me. This hydrocarbon group occurs in many organic compounds. It is a very stable group in most molecules. While the methyl group is usually part of a larger molecule, bounded to the rest of the molecule by a single covalent bond (), it can be found on its own in any of three forms: methanide anion (), methylium cation () or methyl radical (). The anion has eight valence electrons, the radical seven and the cation six. All three forms are highly reactive and rarely observed. Methyl cation, anion, and radical Methyl cation The methylium cation () exists in the gas phase, but is otherwise not encountered. Some compounds are considered to be sources of the cation, and this simplification is used pervasively in organic chemistry. For example, protonation of methanol gives an elect ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Methyltransferase
In biochemistry, the DNA methyltransferase (DNA MTase, DNMT) family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl methionine (SAM) as the methyl donor. Classification Substrate MTases can be divided into three different groups on the basis of the chemical reactions they catalyze: * m6A - those that generate N6-methyladenine * m4C - those that generate N4-methylcytosine * m5C - those that generate C5-methylcytosine m6A and m4C methyltransferases are found primarily in prokaryotes (although recent evidence has suggested that m6A is abundant in eukaryotes). m5C methyltransfereases are found in some lower eukaryotes, in most higher plants, and in animals beginning with the echinoderms. The m6A methyltransferases (N-6 adenine-specific DNA methylase) (A-Mtase) are enzymes that specifically methylate the amino group at the C-6 position of adenine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Synaptic Plasticity
In neuroscience, synaptic plasticity is the ability of synapses to strengthen or weaken over time, in response to increases or decreases in their activity. Since memories are postulated to be represented by vastly interconnected neural circuits in the brain, synaptic plasticity is one of the important neurochemical foundations of learning and memory (''see Hebbian theory''). Plastic change often results from the alteration of the number of neurotransmitter receptors located on a synapse. There are several underlying mechanisms that cooperate to achieve synaptic plasticity, including changes in the quantity of neurotransmitters released into a synapse and changes in how effectively cells respond to those neurotransmitters. Synaptic plasticity in both excitatory and inhibitory synapses has been found to be dependent upon postsynaptic calcium release. Historical discoveries In 1973, Terje Lømo and Tim Bliss first described the now widely studied phenomenon of long-term pote ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
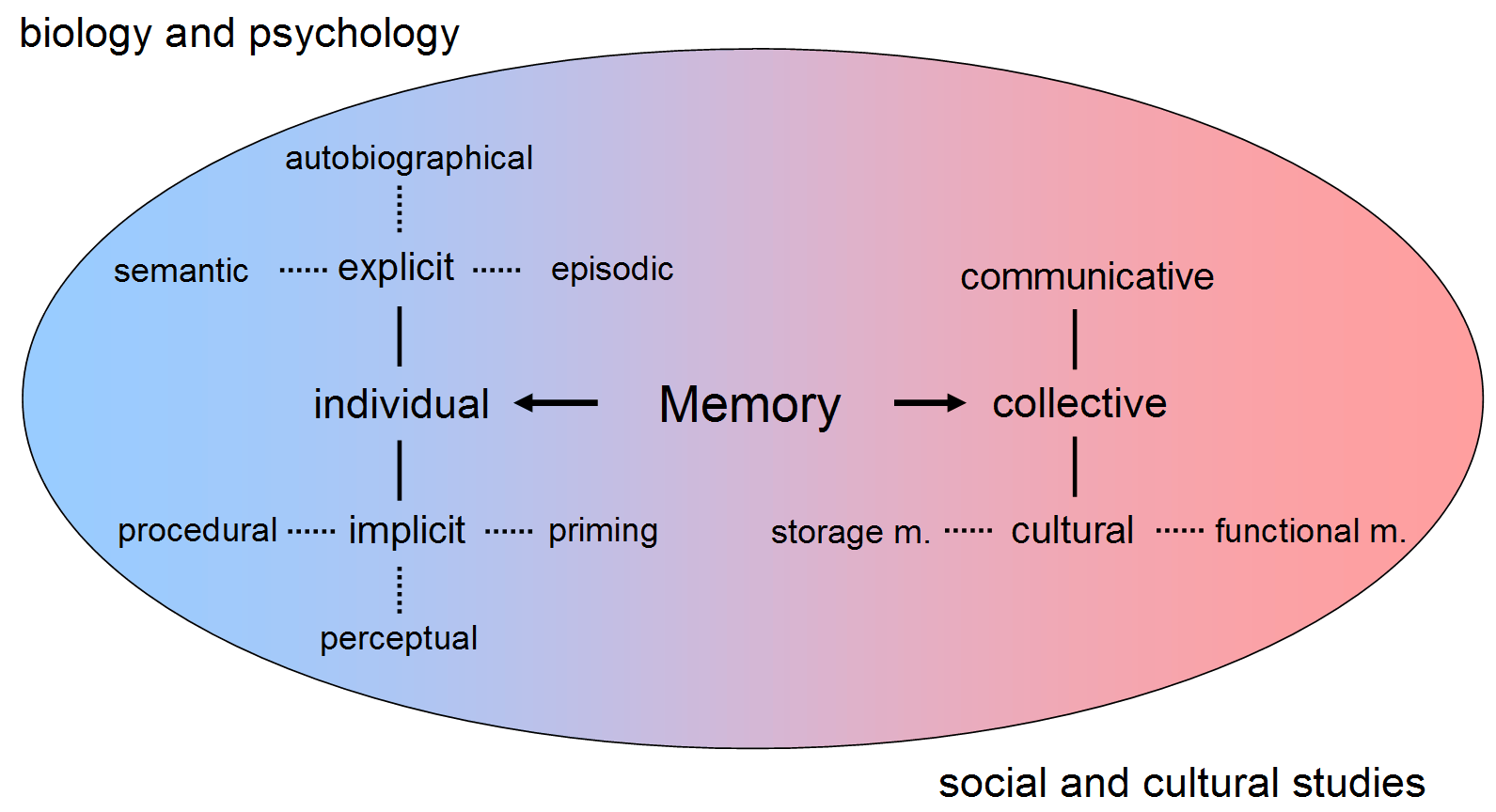
Memory
Memory is the faculty of the mind by which data or information is encoded, stored, and retrieved when needed. It is the retention of information over time for the purpose of influencing future action. If past events could not be remembered, it would be impossible for language, relationships, or personal identity to develop. Memory loss is usually described as forgetfulness or amnesia. Memory is often understood as an informational processing system with explicit and implicit functioning that is made up of a sensory processor, short-term (or working) memory, and long-term memory. This can be related to the neuron. The sensory processor allows information from the outside world to be sensed in the form of chemical and physical stimuli and attended to various levels of focus and intent. Working memory serves as an encoding and retrieval processor. Information in the form of stimuli is encoded in accordance with explicit or implicit functions by the working memory processor. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
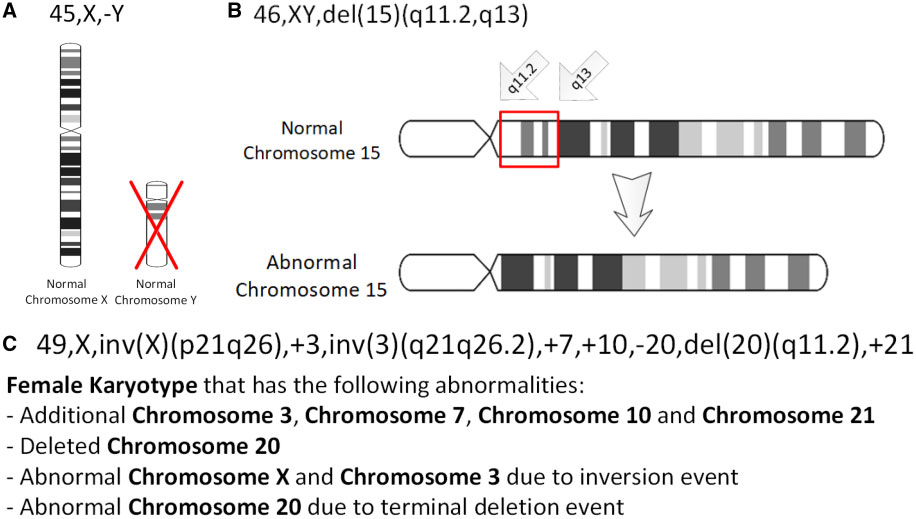
Deletion (genetics)
In genetics, a deletion (also called gene deletion, deficiency, or deletion mutation) (sign: Δ) is a mutation (a genetic aberration) in which a part of a chromosome or a sequence of DNA is left out during DNA replication. Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome. Some chromosomes have fragile spots where breaks occur which result in the deletion of a part of chromosome. The breaks can be induced by heat, viruses, radiations, chemicals. When a chromosome breaks, a part of it is deleted or lost, the missing piece of chromosome is referred to as deletion or a deficiency. For synapsis to occur between a chromosome with a large intercalary deficiency and a normal complete homolog, the unpaired region of the normal homolog must loop out of the linear structure into a deletion or compensation loop. The smallest single base deletion mutations occur by a single base flipping in the template DNA, followed by template DNA strand sli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
_and_with_tumorigenesis_(B)._Brown_shows_8-oxo-dG.jpg)