|
5′ Flanking Region
The 5′ flanking region is a region of DNA adjacent to the 5′ end of the gene. The 5′ flanking region contains the promoter and may contain enhancers or other protein-binding sites. It is the region of DNA that is not transcribed into RNA. This region, not to be confused with the 5′ untranslated region, is not transcribed into RNA or translated into a functional protein. These regions' primary function is the regulation of gene transcription. 5′ flanking regions are categorized between prokaryotes and eukaryotes. Eukaryotic elements In eukaryotes, the 5′ flanking region has complex regulatory elements such as enhancers, silencers, and promoters. The primary promoter element in eukaryotes is the TATA box. Other promoter elements in eukaryotic 5′ flanking regions include initiator elements, downstream core promoter elements, CAAT box, and the GC box. Enhancer Enhancers are DNA sequences found in 5′ flanking regions of eukaryotic genes that affect trans ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Directionality (molecular Biology)
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide Pentose, pentose-sugar-ring means that there will be a 5′ end (usually pronounced "five-prime end"), which frequently contains a phosphate group attached to the 5′ carbon of the ribose ring, and a 3′ end (usually pronounced "three-prime end"), which typically is unmodified from the ribose -OH substituent. In a DNA double helix, the strands run in opposite directions to permit base pairing between them, which is essential for replication or Transcription (biology), transcription of the encoded information. Nucleic acids can only be synthesized in vivo in the 5′-to-3′ direction, as the polymerases that assemble various types of new strands generally rely on the energy produced by breaking nucleoside triphosphate bonds to attach new nucleo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are Gene expression, expressed in the desired Cell (biology), cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are approximately 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ccaat-enhancer-binding Proteins
CCAAT-enhancer-binding proteins (or C/EBPs) is a Protein family, family of transcription factors composed of six members, named from C/EBPα to C/EBPζ. They promote the expression of certain genes through interaction with their Promoter (biology), promoters. Once bound to DNA, C/EBPs can recruit so-called co-activators (such as CBP) that in turn can open up chromatin structure or recruit basal transcription factors. Function C/EBP proteins interact with the CCAAT (cytosine-cytosine-adenosine-adenosine-thymidine) box motif, which is present in several gene promoters. They are characterized by a highly conservation (genetics), conserved bZIP domain, basic-leucine zipper (bZIP) domain at the C-terminus. This domain is involved in protein dimer, dimerization and DNA binding, as are other transcription factors of the leucine zipper domain-containing family (''c-Fos'' and ''c-jun''). The bZIP domain structure of C/EBPs is composed of an α-helix that forms a "coiled coil" structure ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Upstream And Downstream (DNA)
In molecular biology and genetics, upstream and downstream both refer to relative positions of genetic code in DNA or RNA. Each strand of DNA or RNA has a 5' end and a 3' end, so named for the carbon position on the deoxyribose (or ribose) ring. By convention, upstream and downstream relate to the 5' to 3' direction respectively in which RNA transcription takes place. Upstream is toward the 5' end of the RNA molecule, and downstream is toward the 3' end. When considering double-stranded DNA, upstream is toward the 5' end of the coding strand for the gene in question and downstream is toward the 3' end. Due to the anti-parallel nature of DNA, this means the 3' end of the template strand is upstream of the gene and the 5' end is downstream. Some genes on the same DNA molecule may be transcribed in opposite directions. This means the upstream and downstream areas of the molecule may change depending on which gene is used as the reference. The terms upstream and downstream are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleotide
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all Life, life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common Nutrient, nutrients by the liver. Nucleotides are composed of three subunit molecules: a nucleobase, a pentose, five-carbon sugar (ribose or deoxyribose), and a phosphate group consisting of one to three phosphates. The four nucleobases in DNA are guanine, adenine, cytosine, and thymine; in RNA, uracil is used in place of thymine. Nucleotides also play a central role in metabolism at a fundamental, cellular level. They provide chemical energy—in the form of the nucleoside triphosphates, adenosine triphosphate (ATP), guanosine triphosphate (GTP), cytidine triphosphate (CTP), and uridine triph ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
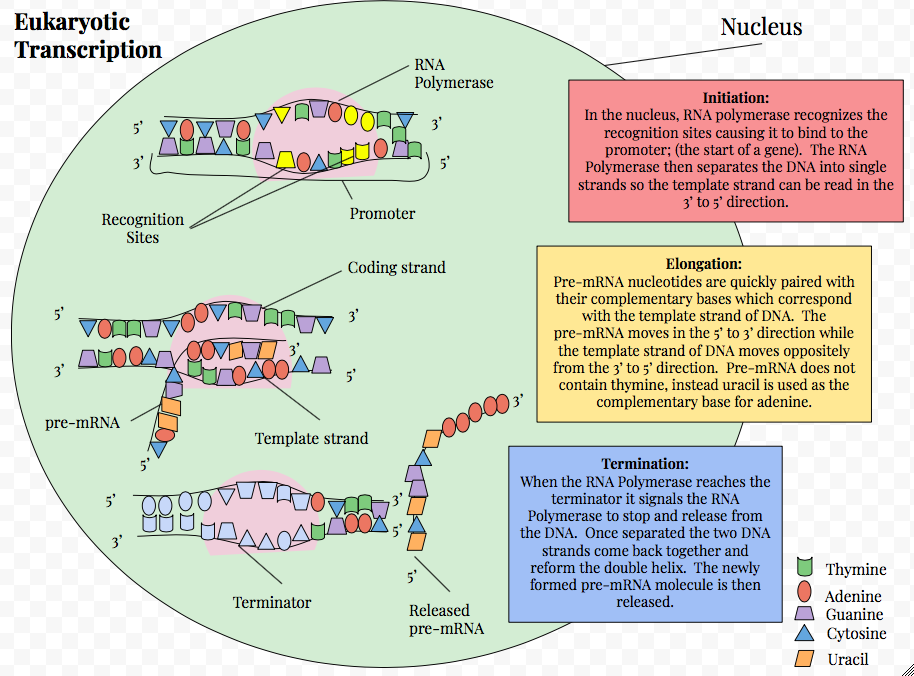
Eukaryotic Transcription
Eukaryotic transcription is the elaborate process that eukaryotic cells use to copy genetic information stored in DNA into units of transportable complementary RNA replica. Gene transcription occurs in both eukaryotic and prokaryotic cells. Unlike prokaryotic RNA polymerase that initiates the transcription of all different types of RNA, RNA polymerase in eukaryotes (including humans) comes in three variations, each translating a different type of gene. A eukaryotic cell has a nucleus that separates the processes of transcription and translation. Eukaryotic transcription occurs within the nucleus where DNA is packaged into nucleosomes and higher order chromatin structures. The complexity of the eukaryotic genome necessitates a great variety and complexity of gene expression control. Eukaryotic transcription proceeds in three sequential stages: initiation, elongation, and termination. The RNAs transcribed serve diverse functions. For example, structural components of the ribo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor II H
Transcription factor II H (TFIIH) is an important protein complex, having roles in transcription of various protein-coding genes and DNA nucleotide excision repair (NER) pathways. TFIIH first came to light in 1989 when general transcription factor-δ or basic transcription factor 2 was characterized as an indispensable transcription factor in vitro. This factor was also isolated from yeast and finally named TFIIH in 1992. TFIIH consists of ten subunits, 7 of which (ERCC2/XPD, ERCC3/XPB, GTF2H1/p62, GTF2H4/p52, GTF2H2/p44, GTF2H3/p34 and GTF2H5/TTDA) form the core complex. The cyclin-activating kinase-subcomplex (CDK7, MAT1, and cyclin H) is linked to the core via the XPD protein. Two of the subunits, ERCC2/XPD and ERCC3/XPB, have helicase and ATPase activities and help create the transcription bubble. In a test tube, these subunits are only required for transcription if the DNA template is not already DNA melting, denatured or if it is DNA supercoil, supercoiled. Two other TFII ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor II F
Transcription factor II F (TFIIF) is one of several general transcription factors that make up the RNA polymerase II preinitiation complex. TFIIF is encoded by the , , and genes. TFIIF binds to RNA polymerase II RNA polymerase II (RNAP II and Pol II) is a Protein complex, multiprotein complex that Transcription (biology), transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNA pol ... when the enzyme is already unbound to any other transcription factor, thus preventing it from contacting DNA outside the promoter. Furthermore, TFIIF stabilizes the RNA polymerase II while it's contacting TBP and TFIIB. See also * TFIIA * TFIIB * TFIID * TFIIE * TFIIH References External links * Helicases Molecular genetics Proteins Gene expression Transcription factors {{genetics-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor II B
Transcription factor II B (TFIIB) is a general transcription factor that is involved in the formation of the RNA polymerase II preinitiation complex (PIC) and aids in stimulating transcription initiation. TFIIB is localised to the nucleus and provides a platform for PIC formation by binding and stabilising the DNA-TBP (TATA-binding protein) complex and by recruiting RNA polymerase II and other transcription factors. It is encoded by the gene, and is homologous to archaeal transcription factor B and analogous to bacterial sigma factors. Structure TFIIB is a single 33kDa polypeptide consisting of 316 amino acids. TFIIB is made up of four functional regions: the C-terminal core domain; the B linker; the B reader and the amino terminal zinc ribbon. TFIIB makes protein-protein interactions with the TATA-binding protein (TBP) subunit of transcription factor IID, and the RPB1 subunit of RNA polymerase II. TFIIB makes sequence-specific protein-DNA interactions with the B rec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor II D
Transcription factor II D (TFIID) is one of several general transcription factors that make up the RNA polymerase II preinitiation complex. RNA polymerase II holoenzyme is a form of eukaryotic RNA polymerase II that is recruited to the promoters of protein-coding genes in living cells. It consists of RNA polymerase II, a subset of general transcription factors, and regulatory proteins known as SRB proteins. Before the start of transcription, the transcription Factor II D (TFIID) complex binds to the core promoter DNA of the gene through specific recognition of promoter sequence motifs, including the TATA box, Initiator, Downstream Promoter, Motif Ten, or Downstream Regulatory elements. Functions * Coordinates the activities of more than 70 polypeptides required for initiation of transcription by RNA polymerase II * Binds to the core promoter to position the polymerase properly * Serves as the scaffold for assembly of the remainder of the transcription complex * Acts as a c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TATA-binding Protein
The TATA-binding protein (TBP) is a general transcription factor that binds to a DNA sequence called the TATA box. This DNA sequence is found about 30 base pairs upstream of the transcription start site in some eukaryotic gene promoters. TBP gene family TBP is a member of a small gene family of TBP-related factors. The first TBP-related factor (TRF/TRF1) was identified in the fruit fly Drosophila, but appears to be fly or insect-specific. Subsequently TBPL1/TRF2 was found in the genomes of many metazoans, whereas vertebrate genomes encode a third vertebrate family member, TBPL2/TRF3. In specific cell types or on specific promoters TBP can be replaced by one of these TBP-related factors, some of which interact with the TATA box similarly to TBP. Role as transcription factor TBP is a subunit of the eukaryotic general transcription factor TFIID. TFIID is the first protein to bind to DNA during the formation of the transcription preinitiation complex of RNA polymerase ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase II
RNA polymerase II (RNAP II and Pol II) is a Protein complex, multiprotein complex that Transcription (biology), transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNA polymerase, RNAP enzymes found in the nucleus of eukaryote, eukaryotic cells. A 550 kDa complex of 12 subunits, RNAP II is the most studied type of RNA polymerase. A wide range of transcription factors are required for it to bind to upstream gene promoter (biology), promoters and begin transcription. Discovery Early studies suggested a minimum of two RNAPs: one which synthesized rRNA in the nucleolus, and one which synthesized other RNA in the nucleoplasm, part of the nucleus but outside the nucleolus. In 1969, biochemists Robert G. Roeder and William J. Rutter, William Rutter discovered there are total three distinct nuclear RNA polymerases, an additional RNAP that was responsible for transcription of some kind of RNA in the nucleoplasm. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |