|
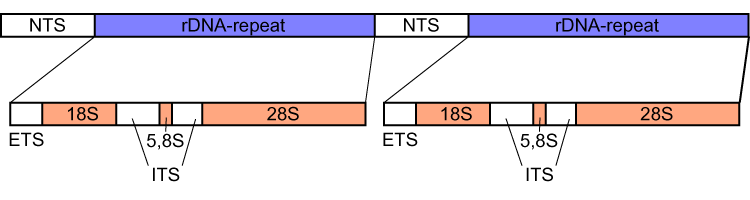
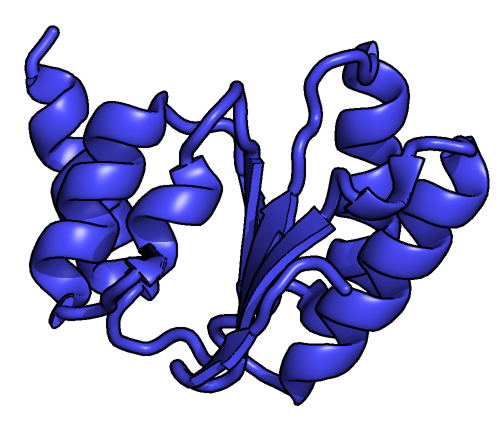
5.8S RRNA
In molecular biology, the 5.8S ribosomal RNA (5.8S rRNA) is a non-coding RNA component of the large subunit of the Eukaryote, eukaryotic ribosome and so plays an important role in translation (genetics), protein translation. It is Transcription (genetics), transcribed by RNA polymerase I as part of the 45S precursor that also contains 18S rRNA, 18S and 28S rRNA. Its function is thought to be in ribosome translocation. It is also known to form covalent linkage to the p53 tumour suppressor protein. 5.8S rRNA can be used as a reference gene for miRNA detection. The 5.8S ribosomal RNA is used to better understand other rRNA processes and pathways in the cell. The 5.8S rRNA is homologous to the 5' end of non-eukaryotic LSU rRNA. In eukaryotes, the insertion of ITS2 breaks LSU rRNA into 5.8S and 28S rRNAs. Some Diptera, flies have their 5.8 rRNA further split into two pieces. Structure L567.5 rRNA structure is approximately 150 nucleotides in size and it consists of plenty of folded ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
28S RRNA
Crazy Eights is a shedding-type card game for two to seven players and the best known American member of the Eights Group which also includes Pig and Spoons. The object of the game is to be the first player to discard all of their cards. The game is similar to Switch and Mau Mau. Originally this was played primarily by children with the left over cards not used in Euchre. Now a standard 52-card deck is used when there are five or fewer players. When there are more than five players, two decks are shuffled together and all 104 cards are used. Origins The game first appeared as ''Eights'' in the 1930s, and the name ''Crazy Eights'' dates to the 1940s, derived from the United States military designation for discharge of mentally unstable soldiers, Section 8. It may have derived from the German game of Mau-Mau. There are many variations of the basic game, under names including ''Craits'', '' Last Card'', ''Switch'', and ''Black Jack''. Bartok, Mao, Taki, and Uno add further ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endonuclease
Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (without regard to sequence), while many, typically called restriction endonucleases or restriction enzymes, cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (endo) portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity. Restriction enzymes are endonucleases from eubacteria and archaea that recognize a specific DNA sequence. The nucleotide sequence recognized for cleavage by a restriction enzyme is called the restriction site. Typically, a restriction site will be a palindromic sequence about four to six nucleotides ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
External Transcribed Spacer
External transcribed spacer (ETS) refers to a piece of non-functional RNA, closely related to the internal transcribed spacer, which is situated outside structural ribosomal RNAs (rRNA) on a common precursor transcript. ETS sequences characteristic to an organism can be used to trace its phylogeny A phylogenetic tree (also phylogeny or evolutionary tree Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA.) is a branching diagram or a tree showing the evolutionary relationships among various biological spec .... References {{genetics-stub DNA Phylogenetics de:Spacer (Biologie) ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Internal Transcribed Spacer
Internal transcribed spacer (ITS) is the spacer DNA situated between the small-subunit ribosomal RNA (rRNA) and large-subunit rRNA genes in the chromosome or the corresponding transcribed region in the polycistronic rRNA precursor transcript. ITS across life domains In bacteria and archaea, there is a single ITS, located between the 16S and 23S rRNA genes. Conversely, there are two ITSs in eukaryotes: ITS1 is located between 18S and 5.8S rRNA genes, while ITS2 is between 5.8S and 28S (in opisthokonts, or 25S in plants) rRNA genes. ITS1 corresponds to the ITS in bacteria and archaea, while ITS2 originated as an insertion that interrupted the ancestral 23S rRNA gene. Organization In bacteria and archaea, the ITS occurs in one to several copies, as do the flanking 16S and 23S genes. When there are multiple copies, these do not occur adjacent to one another. Rather, they occur in discrete locations in the circular chromosome. It is not uncommon in bacteria to carry tRN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryotic Large Ribosomal Subunit (60S)
Ribosomal particles are denoted according to their sedimentation coefficients in Svedberg units. The 60S subunit is the large subunit of eukaryotic 80S ribosomes. It is structurally and functionally related to the 50S subunit of 70S prokaryotic ribosomes. However, the 60S subunit is much larger than the prokaryotic 50S subunit and contains many additional protein segments, as well as ribosomal RNA expansion segments. Overall structure Characteristic features of the large subunit, shown below in the "Crown View", include the central protuberance (CP) and the two stalks, which are named according to their bacterial protein components (L1 stalk on the left as seen from the subunit interface and L7/L12 on the right). There are three binding sites for tRNA, the A-site, P-site and E-site (see article on protein translation for details). The core of the 60S subunit is formed by the 28S ribosomal RNA (abbreviated 28S rRNA), which is homologous to the prokaryotic 23S rRNA, which also co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosomal Proteins
A ribosomal protein (r-protein or rProtein) is any of the proteins that, in conjunction with rRNA, make up the ribosomal subunits involved in the cellular process of translation. ''E. coli'', other bacteria and Archaea have a 30S small subunit and a 50S large subunit, whereas humans and yeasts have a 40S small subunit and a 60S large subunit. Equivalent subunits are frequently numbered differently between bacteria, Archaea, yeasts and humans. A large part of the knowledge about these organic molecules has come from the study of '' E. coli'' ribosomes. All ribosomal proteins have been isolated and many specific antibodies have been produced. These, together with electronic microscopy and the use of certain reactives, have allowed for the determination of the topography of the proteins in the ribosome. More recently, a near-complete (near)atomic picture of the ribosomal proteins is emerging from the latest high-resolution cryo-EM data (including ). Conservation Ribosomal prot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleotides
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver. Nucleotides are composed of three subunit molecules: a nucleobase, a five-carbon sugar (ribose or deoxyribose), and a phosphate group consisting of one to three phosphates. The four nucleobases in DNA are guanine, adenine, cytosine and thymine; in RNA, uracil is used in place of thymine. Nucleotides also play a central role in metabolism at a fundamental, cellular level. They provide chemical energy—in the form of the nucleoside triphosphates, adenosine triphosphate (ATP), guanosine triphosphate (GTP), cytidine triphosphate (CTP) and uridine triphosphate (UTP)—throughout the cell for the many cellular fun ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Diptera
Flies are insects of the order Diptera, the name being derived from the Greek δι- ''di-'' "two", and πτερόν ''pteron'' "wing". Insects of this order use only a single pair of wings to fly, the hindwings having evolved into advanced mechanosensory organs known as halteres, which act as high-speed sensors of rotational movement and allow dipterans to perform advanced aerobatics. Diptera is a large order containing an estimated 1,000,000 species including horse-flies, crane flies, hoverflies and others, although only about 125,000 species have been described. Flies have a mobile head, with a pair of large compound eyes, and mouthparts designed for piercing and sucking (mosquitoes, black flies and robber flies), or for lapping and sucking in the other groups. Their wing arrangement gives them great maneuverability in flight, and claws and pads on their feet enable them to cling to smooth surfaces. Flies undergo complete metamorphosis; the eggs are often laid on the l ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
LSU RRNA
Large subunit ribosomal ribonucleic acid (LSU rRNA) is the largest of the two major RNA components of the ribosome. Associated with a number of ribosomal proteins, the LSU rRNA forms the large subunit of the ribosome. The LSU rRNA acts as a ribozyme, catalyzing Peptidyl transferase, peptide bond formation. Characteristics Use in phylogenetics LSU rRNA sequences are widely used for working out Phylogenetics, evolutionary relationships among organisms, since they are of ancient origin and are found in all known forms of life. See also *SSU rRNA: the small subunit ribosomal ribonucleic acid. References {{ribosome subunits Ribosomal RNA Protein biosynthesis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MiRNA
MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides. Found in plants, animals and some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRNAs base-pair to complementary sequences in mRNA molecules, then gene silence said mRNA molecules by one or more of the following processes: (1) cleavage of mRNA strand into two pieces, (2) destabilization of mRNA by shortening its poly(A) tail, or (3) translation of mRNA into proteins. This last method of gene silencing is the least efficient of the three, and requires the aid of ribosomes. miRNAs resemble the small interfering RNAs (siRNAs) of the RNA interference (RNAi) pathway, except miRNAs derive from regions of RNA transcripts that fold back on themselves to form short hairpins, whereas siRNAs derive from longer regions of double-stranded RNA. The human genome may encode over 1900 miRNAs, although more recent analysis suggests that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Covalent
A covalent bond is a chemical bond that involves the sharing of electrons to form electron pairs between atoms. These electron pairs are known as shared pairs or bonding pairs. The stable balance of attractive and repulsive forces between atoms, when they share electrons, is known as covalent bonding. For many molecules, the sharing of electrons allows each atom to attain the equivalent of a full valence shell, corresponding to a stable electronic configuration. In organic chemistry, covalent bonding is much more common than ionic bonding. Covalent bonding also includes many kinds of interactions, including σ-bonding, π-bonding, metal-to-metal bonding, agostic interactions, bent bonds, three-center two-electron bonds and three-center four-electron bonds. The term ''covalent bond'' dates from 1939. The prefix ''co-'' means ''jointly, associated in action, partnered to a lesser degree, '' etc.; thus a "co-valent bond", in essence, means that the atoms share " valence", such a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |

_(10144905255).jpg)