|
23S RRNA (adenine2085-N6)-dimethyltransferase
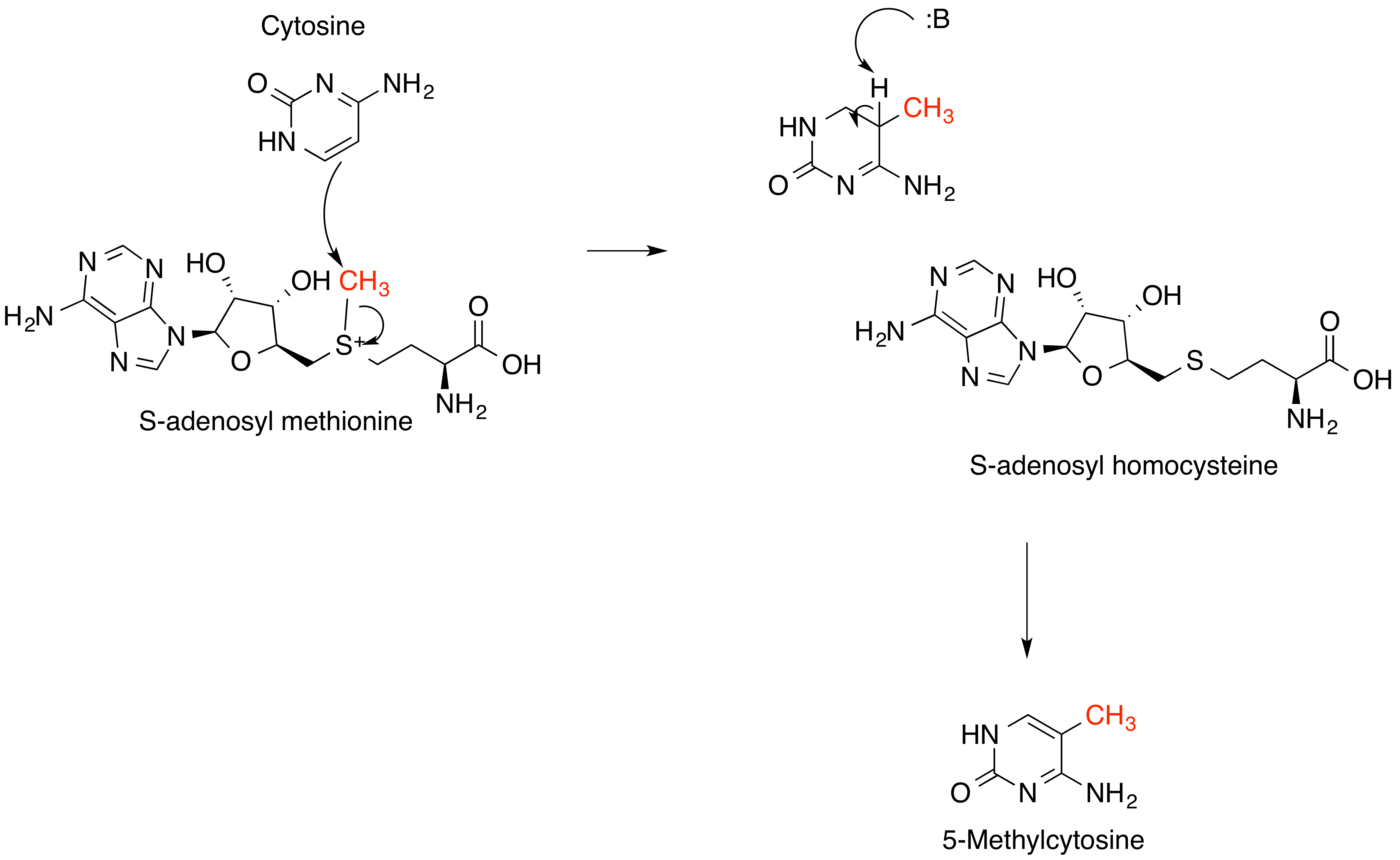
23S rRNA (adenine2085-N6)-dimethyltransferase (, ''ErmC' methyltransferase, ermC methylase, ermC 23S rRNA methyltransferase, rRNA:m6A methyltransferase ErmC', ErmC', rRNA methyltransferase ErmC' '') is an enzyme with systematic name ''S-adenosyl-L-methionine:23S rRNA (adenine2085-N6)-dimethyltransferase''. This enzyme catalyses the following chemical reaction : 2 S-adenosyl-L-methionine + adenine2085 in 23S rRNA \rightleftharpoons 2 S-adenosyl-L-homocysteine + N6- dimethyladenine2085 in 23S rRNA ErmC is a methyltransferase that confers resistance to the macrolide- lincosamide-streptogramin B group of antibiotics by catalysing the methylation In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These t ... of 23S rRNA at adenine2085. References External links * {{Portal bar, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Enzymes
This article lists enzymes by their classification in the International Union of Biochemistry and Molecular Biology's Enzyme Commission (EC) numbering system. * List of EC numbers (EC 5) * List of EC numbers (EC 6) :Oxidoreductases (EC 1) (Oxidoreductase) *Dehydrogenase * Luciferase *DMSO reductase :EC 1.1 (act on the CH-OH group of donors) * :EC 1.1.1 (with NAD+ or NADP+ as acceptor) ** Alcohol dehydrogenase (NAD) ** Alcohol dehydrogenase (NADP) **Homoserine dehydrogenase ** Aminopropanol oxidoreductase **Diacetyl reductase **Glycerol dehydrogenase **Propanediol-phosphate dehydrogenase ** glycerol-3-phosphate dehydrogenase (NAD+) ** D-xylulose reductase **L-xylulose reductase **Lactate dehydrogenase **Malate dehydrogenase **Isocitrate dehydrogenase ** HMG-CoA reductase * :EC 1.1.2 (with a cytochrome as acceptor) * :EC 1.1.3 (with oxygen as acceptor) **Glucose oxidase **L-gulonolactone oxidase **Thiamine oxidase **Xanthine oxidase * :EC 1.1.4 (with a disul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Catalysis
Catalysis () is the process of increasing the rate of a chemical reaction by adding a substance known as a catalyst (). Catalysts are not consumed in the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recycles quickly, very small amounts of catalyst often suffice; mixing, surface area, and temperature are important factors in reaction rate. Catalysts generally react with one or more reactants to form intermediates that subsequently give the final reaction product, in the process of regenerating the catalyst. Catalysis may be classified as either homogeneous, whose components are dispersed in the same phase (usually gaseous or liquid) as the reactant, or heterogeneous, whose components are not in the same phase. Enzymes and other biocatalysts are often considered as a third category. Catalysis is ubiquitous in chemical industry of all kinds. Estimates are that 90% of all commercially produced chemical products involve catalysts at some s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemical Reaction
A chemical reaction is a process that leads to the IUPAC nomenclature for organic transformations, chemical transformation of one set of chemical substances to another. Classically, chemical reactions encompass changes that only involve the positions of electrons in the forming and breaking of chemical bonds between atoms, with no change to the Atomic nucleus, nuclei (no change to the elements present), and can often be described by a chemical equation. Nuclear chemistry is a sub-discipline of chemistry that involves the chemical reactions of unstable and radioactive Chemical element, elements where both electronic and nuclear changes can occur. The substance (or substances) initially involved in a chemical reaction are called reagent, reactants or reagents. Chemical reactions are usually characterized by a chemical change, and they yield one or more Product (chemistry), products, which usually have properties different from the reactants. Reactions often consist of a sequence o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
S-adenosyl-L-methionine
''S''-Adenosyl methionine (SAM), also known under the commercial names of SAMe, SAM-e, or AdoMet, is a common cosubstrate involved in methyl group transfers, transsulfuration, and aminopropylation. Although these anabolic reactions occur throughout the body, most SAM is produced and consumed in the liver. More than 40 methyl transfers from SAM are known, to various substrates such as nucleic acids, proteins, lipids and secondary metabolites. It is made from adenosine triphosphate (ATP) and methionine by methionine adenosyltransferase. SAM was first discovered by Giulio Cantoni in 1952. In bacteria, SAM is bound by the SAM riboswitch, which regulates genes involved in methionine or cysteine biosynthesis. In eukaryotic cells, SAM serves as a regulator of a variety of processes including DNA, tRNA, and rRNA methylation; immune response; amino acid metabolism; transsulfuration; and more. In plants, SAM is crucial to the biosynthesis of ethylene, an important plant hormone and sign ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenine
Adenine () ( symbol A or Ade) is a nucleobase (a purine derivative). It is one of the four nucleobases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The three others are guanine, cytosine and thymine. Its derivatives have a variety of roles in biochemistry including cellular respiration, in the form of both the energy-rich adenosine triphosphate (ATP) and the cofactors nicotinamide adenine dinucleotide (NAD), flavin adenine dinucleotide (FAD) and Coenzyme A. It also has functions in protein synthesis and as a chemical component of DNA and RNA. The shape of adenine is complementary to either thymine in DNA or uracil in RNA. The adjacent image shows pure adenine, as an independent molecule. When connected into DNA, a covalent bond is formed between deoxyribose sugar and the bottom left nitrogen (thereby removing the existing hydrogen atom). The remaining structure is called an ''adenine residue'', as part of a larger molecule. Adenosine is ad ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosomal RNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form SSU rRNA, small and LSU rRNA, large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and Translation (biology), translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins by mass. Structure Although the primary structure of rRNA sequences can vary across organisms, Base pair, base-pairing within these sequences commonly forms stem-loop configurations. The length and position of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methyltransferase
Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding ''S''-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the leavi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Macrolide
The Macrolides are a class of natural products that consist of a large macrocyclic lactone ring to which one or more deoxy sugars, usually cladinose and desosamine, may be attached. The lactone rings are usually 14-, 15-, or 16-membered. Macrolides belong to the polyketide class of natural products. Some macrolides have antibiotic or antifungal activity and are used as pharmaceutical drugs. Rapamycin is also a macrolide and was originally developed as an antifungal, but is now used as an immunosuppressant drug and is being investigated as a potential longevity therapeutic. Macrolides are bacteriostatic in that they suppress or inhibit bacterial growth rather than killing bacteria completely. Definition In general, any macrocyclic lactone having greater than 8-membered rings are candidates for this class. The macrocycle may contain amino nitrogen, amide nitrogen (but should be differentiated from cyclopeptides), an oxazole ring, or a thiazole ring. Benzene rings are exclude ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lincosamide
Lincosamides are a class of antibiotics, which include lincomycin, clindamycin, and pirlimycin. Structure Lincosamides consist of a pyrrolidine ring linked to a pyranose moiety (methylthio-lincosamide) via an amide bond. Hydrolysis of lincosamides, specifically lincomycin, splits the molecule into its building blocks of the sugar and proline moieties. Both of these derivatives can conversely be recombined into the drug itself or a derivative. Synthesis Biosynthesis of lincosamides occurs through a biphasic pathway, in which propylproline and methylthiolincosamide are independently synthesized immediately before condensation of the two precursor molecules. Condensation of the propylproline carboxyl group with the methylthiolincosamide amine group via an amide bond forms ''N''-demethyllincomycin. ''N''-Demethyllincomycin is subsequently methylated via ''S''-adenosyl methionine to produce lincomycin A. Lincomycin is naturally produced by bacteria species, namely ''Streptomy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |