|
(myosin-light-chain) Phosphatase
Myosin light-chain phosphatase, also called myosin phosphatase (EC 3.1.3.53; systematic name yosin-light-chainphosphate phosphohydrolase), is an enzyme (specifically a serine/threonine-specific protein phosphatase) that dephosphorylates the regulatory light chain of myosin II: : yosin light-chainphosphate + H2O = yosin light-chain+ phosphate This dephosphorylation reaction occurs in smooth muscle tissue and initiates the relaxation process of the muscle cells. Thus, myosin phosphatase undoes the muscle contraction process initiated by myosin light-chain kinase. The enzyme is composed of three subunits: the catalytic region ( protein phosphatase 1, or PP1), the myosin binding subunit (MYPT1), and a third subunit (M20) of unknown function. The catalytic region uses two manganese ions as catalysts to dephosphorylate the light-chains on myosin, which causes a conformational change in the myosin and relaxes the muscle. The enzyme is highly conserved and is found in all ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Crossbridge
The sliding filament theory explains the mechanism of muscle contraction based on muscle proteins that slide past each other to generate movement. According to the sliding filament theory, the myosin (thick filaments) of muscle fibers slide past the actin (thin filaments) during muscle contraction, while the two groups of filaments remain at relatively constant length. The theory was independently introduced in 1954 by two research teams, one consisting of Andrew Huxley and Rolf Niedergerke from the University of Cambridge, and the other consisting of Hugh Huxley and Jean Hanson from the Massachusetts Institute of Technology. It was originally conceived by Hugh Huxley in 1953. Andrew Huxley and Niedergerke introduced it as a "very attractive" hypothesis. Before the 1950s there were several competing theories on muscle contraction, including electrical attraction, protein folding, and protein modification. The novel theory directly introduced a new concept called cross-bridge th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme Inhibitor
An enzyme inhibitor is a molecule that binds to an enzyme and blocks its activity. Enzymes are proteins that speed up chemical reactions necessary for life, in which substrate molecules are converted into products. An enzyme facilitates a specific chemical reaction by binding the substrate to its active site, a specialized area on the enzyme that accelerates the most difficult step of the reaction. An enzyme inhibitor stops ("inhibits") this process, either by binding to the enzyme's active site (thus preventing the substrate itself from binding) or by binding to another site on the enzyme such that the enzyme's catalysis of the reaction is blocked. Enzyme inhibitors may bind reversibly or irreversibly. Irreversible inhibitors form a chemical bond with the enzyme such that the enzyme is inhibited until the chemical bond is broken. By contrast, reversible inhibitors bind non-covalently and may spontaneously leave the enzyme, allowing the enzyme to resume its function. Reve ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Telokin
Telokin (also known as kinase-related protein or KRP) is an abundant protein found in smooth-muscle. It is identical to the C-terminus of myosin light-chain kinase. Telokin may play a role in the stabilization of unphosphorylated smooth-muscle myosin filaments. Because of its origin as the C-terminal end of smooth muscle myosin light chain kinase, it is called "telokin" (from a combination of the Greek telos, "end" and kinase). Nomenclature and classification Telokin's systematic name is ATP: yosin light chainO-phosphotransferase and its recommended name is myosin-light-chain kinase. (). The gene ''MYLK'', a muscle member of the immunoglobulin gene superfamily, encodes myosin light chain kinase which is a calcium/calmodulin dependent enzyme. Four transcript variants that produce four isoforms of the calcium/calmodulin dependent enzyme have been identified as well as two transcripts that produce two isoforms of telokin. The two transcripts that produce the two telokin isof ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ROCK1
ROCK1 is a protein serine/threonine kinase also known as rho-associated, coiled-coil-containing protein kinase 1. Other common names are ROKβ and P160ROCK. ROCK1 is a major downstream effecter of the small GTPase RhoA and is a regulator of the actomyosin cytoskeleton which promotes contractile force generation. ROCK1 plays a role in cancer and in particular cell motility, metastasis, and angiogenesis. Gene and expression ROCK1 is also the name of the gene that encodes the protein ROCK1, a serine/threonine kinase. ROCK1 is activated when bound to the GTP-bound form of RhoA. The human ROCK1 gene is located on human chromosome 18 with specific location of 18q11.1. The location of the base pair starts at 18,529,703 and ends at 18,691,812 bp and translates into 1354 amino acids. ROCK1 has a ubiquitous tissue distribution, but subcellularly it is thought to colocalize with the centrosomes. This is consistent with its function as a key modulator of cell motility, tumor cell inva ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RhoA
Transforming protein RhoA, also known as Ras homolog family member A (RhoA), is a small GTPase protein in the Rho family of GTPases that in humans is encoded by the ''RHOA'' gene. While the effects of RhoA activity are not all well known, it is primarily associated with cytoskeleton regulation, mostly actin stress fibers formation and actomyosin contractility. It acts upon several effectors. Among them, ROCK1 (Rho-associated, coiled-coil containing protein kinase 1) and DIAPH1 (Diaphanous Homologue 1, a.k.a. hDia1, homologue to mDia1 in mouse, diaphanous in ''Drosophila'') are the best described. RhoA, and the other Rho GTPases, are part of a larger family of related proteins known as the Ras superfamily, a family of proteins involved in the regulation and timing of cell division. RhoA is one of the oldest Rho GTPases, with homologues present in the genomes since 1.5 billion years. As a consequence, RhoA is somehow involved in many cellular processes which emerged throughout evol ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
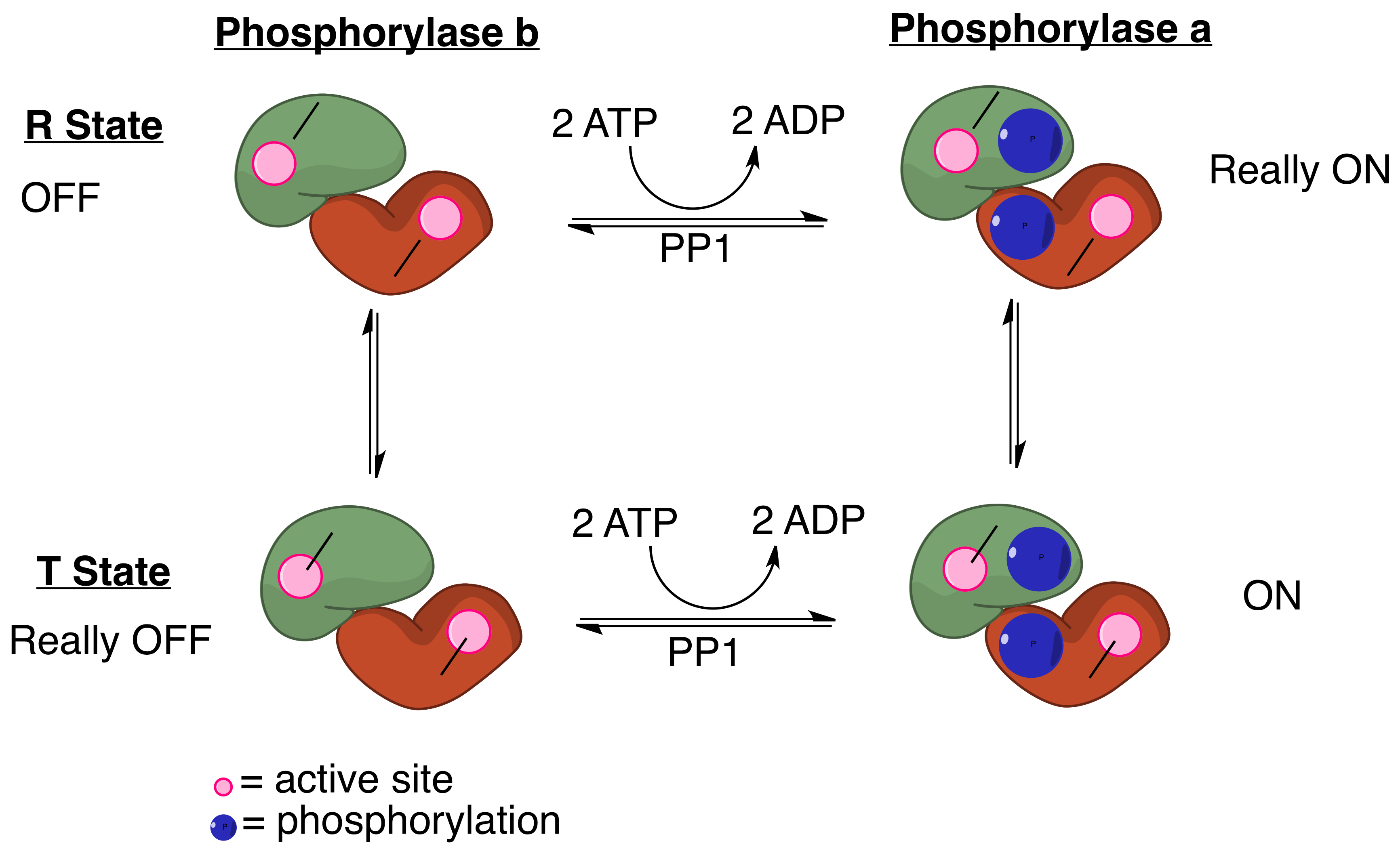
PP1 Mechanism For Myosin Phosphatase
Protein phosphatase 1 (PP1) belongs to a certain class of phosphatases known as protein serine/threonine phosphatases. This type of phosphatase includes metal-dependent protein phosphatases (PPMs) and aspartate-based phosphatases. PP1 has been found to be important in the control of glycogen metabolism, muscle contraction, cell progression, neuronal activities, splicing of RNA, mitosis, cell division, apoptosis, protein synthesis, and regulation of membrane receptors and channels. Structure Each PP1 enzyme contains both a catalytic subunit and at least one regulatory subunit. The catalytic subunit consists of a 30-kD single-domain protein that can form complexes with other regulatory subunits. The catalytic subunit is highly conserved among all eukaryotes, thus suggesting a common catalytic mechanism. The catalytic subunit can form complexes with various regulatory subunits. These regulatory subunits play an important role in substrate specificity as well as compartmentaliz ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |